
py2dmol.solab.org
Integration with AlphaFoldDB (will auto fetch results). Drag and drop results from AF3-server or ColabFold for interactive experience! (1/4)

py2dmol.solab.org
Integration with AlphaFoldDB (will auto fetch results). Drag and drop results from AF3-server or ColabFold for interactive experience! (1/4)
Models for DNA have evolved along separate paths: sequence-to-function (AlphaGenome), language models (Evo2), and generative models (DDSM).
Can these be unified under a single paradigm? 1/15

Models for DNA have evolved along separate paths: sequence-to-function (AlphaGenome), language models (Evo2), and generative models (DDSM).
Can these be unified under a single paradigm? 1/15
www.nature.com/articles/s41...

www.nature.com/articles/s41...

Corrects Tn5 bias in ATAC-seq; improves footprinting, finds elements & reduces false positives.




Corrects Tn5 bias in ATAC-seq; improves footprinting, finds elements & reduces false positives.
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Cerebellum gene reg. driven by TEs, w/cell-type & sp.-specific expression, esp. primate HERVL in granule cells.




Cerebellum gene reg. driven by TEs, w/cell-type & sp.-specific expression, esp. primate HERVL in granule cells.
It was really interesting to write a short manuscript mixing method presentation & broader insights on data preprocessing. 📝
Thank you to my co-supervisors @cantinilab.bsky.social & @juliosaezrod.bsky.social for their feedback on this side work! 🫶
5/5
It was really interesting to write a short manuscript mixing method presentation & broader insights on data preprocessing. 📝
Thank you to my co-supervisors @cantinilab.bsky.social & @juliosaezrod.bsky.social for their feedback on this side work! 🫶
5/5
AlphaFold3, by integrating NMR, X-ray, or cryo-EM data, can generate accurate protein conformation ensembles reflecting experimental conditions.




AlphaFold3, by integrating NMR, X-ray, or cryo-EM data, can generate accurate protein conformation ensembles reflecting experimental conditions.

Excited to share SEAM: Systematic Explanation of Attribtuion-based Mechanisms. SEAM is an explainable AI method that dissects cis-regulatory mechanisms learned by seq2fun genomic deep learning models.
Led by @EESetiz
1/N 🧵👇

Excited to share SEAM: Systematic Explanation of Attribtuion-based Mechanisms. SEAM is an explainable AI method that dissects cis-regulatory mechanisms learned by seq2fun genomic deep learning models.
Led by @EESetiz
1/N 🧵👇
The E2G Portal! e2g.stanford.edu
This collates our predictions of enhancer-gene regulatory interactions across >1,600 cell types and tissues.
Uses cases 👇
1/
The E2G Portal! e2g.stanford.edu
This collates our predictions of enhancer-gene regulatory interactions across >1,600 cell types and tissues.
Uses cases 👇
1/
doi.org/10.48550/arX...
doi.org/10.48550/arX...

doi.org/10.48550/arX...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Corgi imitates cellular gene regulation and integrates DNA sequence and trans-regulator information.
This allows Corgi to make accurate predictions in unseen cell types. Also, it can simulate trans-regulator perturbations in silico.

Corgi imitates cellular gene regulation and integrates DNA sequence and trans-regulator information.
This allows Corgi to make accurate predictions in unseen cell types. Also, it can simulate trans-regulator perturbations in silico.


Hi-C jointly decmp. samples: captures cell states, improves gene pred.




Hi-C jointly decmp. samples: captures cell states, improves gene pred.
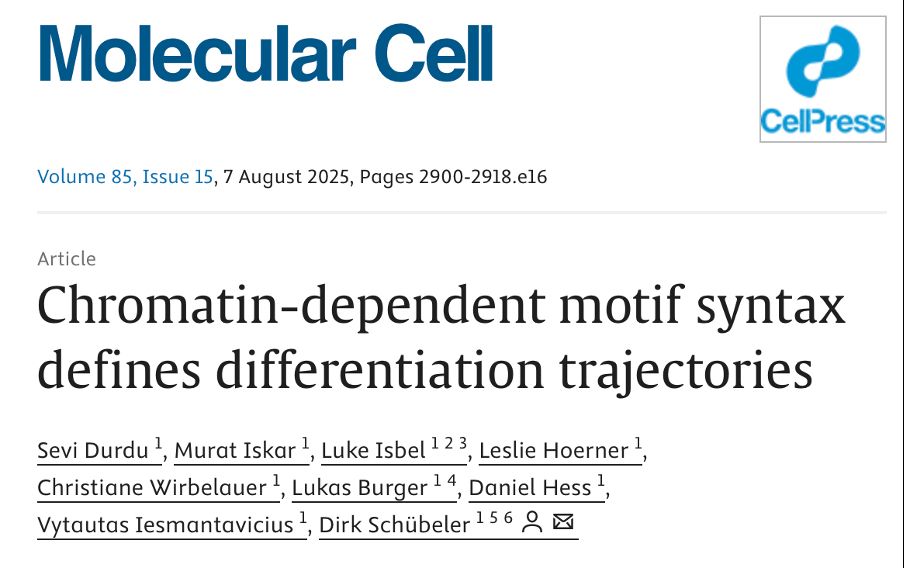
Check this: A scar-free strategy enables seamless editing of multi MB regions without residual sequences
Check this: A scar-free strategy enables seamless editing of multi MB regions without residual sequences
Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵

Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵
www.nature.com/articles/s41...
We show that none of the available* models outperform simple linear baselines. Since the original preprint, we added more methods, metrics, and prettier figures!
🧵


www.nature.com/articles/s41...
We show that none of the available* models outperform simple linear baselines. Since the original preprint, we added more methods, metrics, and prettier figures!
🧵
has an Independent faculty position. Fantastic place to set up your lab –great package: core funding, fantastic Ph.D. students, cutting edge core facilities & great colleagues. Closing date Sept 19th
embl.wd103.myworkdayjobs.com/en-US/EMBL/j...

has an Independent faculty position. Fantastic place to set up your lab –great package: core funding, fantastic Ph.D. students, cutting edge core facilities & great colleagues. Closing date Sept 19th
embl.wd103.myworkdayjobs.com/en-US/EMBL/j...
#Technology #Reproducibility #CellCycle #CellDivision #Statistics
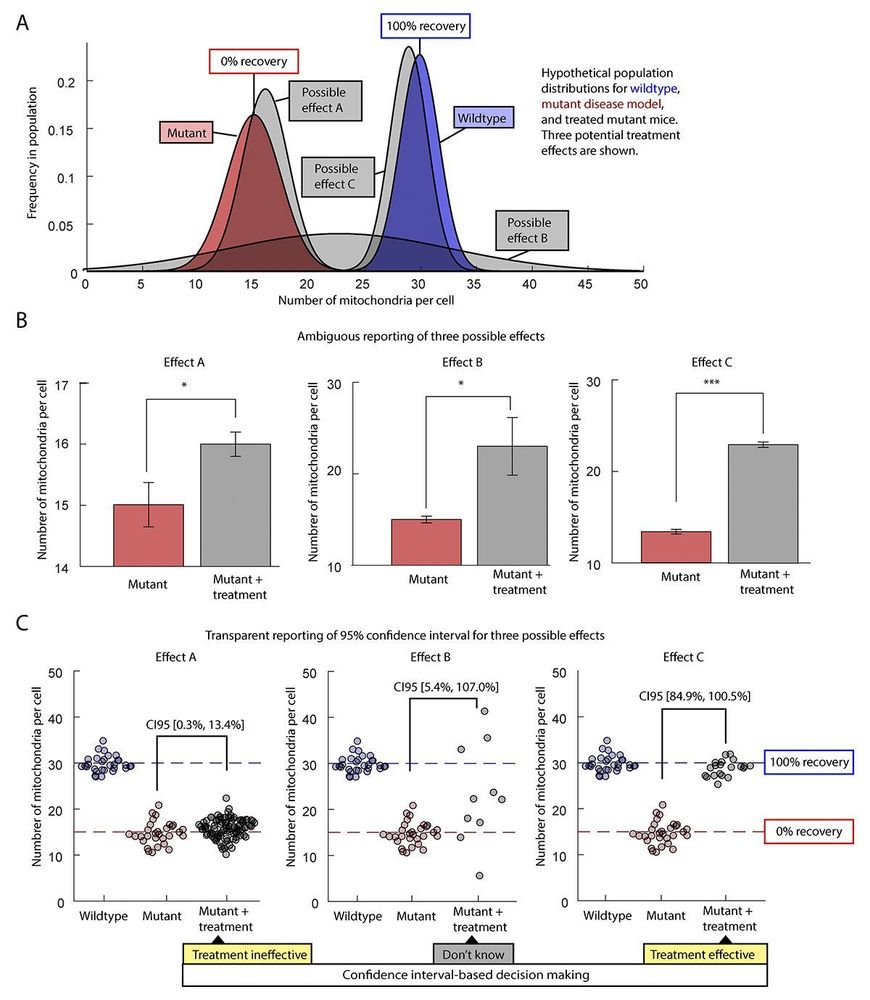
#Technology #Reproducibility #CellCycle #CellDivision #Statistics
Join us, if RNP assembly on centromeres, RNA modifications, & bioinformatics are your thing.
More info also on modiclab.org.
The project is in collaboration with my new collegue Miha Modic. If you are interesed in RNA modification, centromeres and bioinformatics this is for you!
Miha has more positions to fill, infos on his linkedin: linkedin.com/in/miha-modic-9aa8a752.

Join us, if RNP assembly on centromeres, RNA modifications, & bioinformatics are your thing.
More info also on modiclab.org.

