www.biorxiv.org/content/10.6...
The Jan 31st deadline for our FEBS advanced course on integrative structural biology is getting closer, so get your applications in for Lost In Integrative Vol. 2
probingbiomolecules2026.febsevents.org
network.febs.org/posts/integr...

The Jan 31st deadline for our FEBS advanced course on integrative structural biology is getting closer, so get your applications in for Lost In Integrative Vol. 2
probingbiomolecules2026.febsevents.org
network.febs.org/posts/integr...
Our review on computational design of intrinsically disordered proteins is now published in final form 🍝
With @giuliotesei.bsky.social & @fpesce.bsky.social
doi.org/10.1016/j.sb...

Our review on computational design of intrinsically disordered proteins is now published in final form 🍝
With @giuliotesei.bsky.social & @fpesce.bsky.social
doi.org/10.1016/j.sb...
Extending Conformational Ensemble Prediction to Multidomain Proteins and Protein Complex
doi.org/10.64898/202...

Extending Conformational Ensemble Prediction to Multidomain Proteins and Protein Complex
doi.org/10.64898/202...

See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...

See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
Join us in beautiful Aarhus for a 2-day workshop on biomolecular simulations!
9-10 December 2025
Link for registration: aias.au.dk/events/show/...
Join us in beautiful Aarhus for a 2-day workshop on biomolecular simulations!
9-10 December 2025
Link for registration: aias.au.dk/events/show/...
@dipcehu.bsky.social
www.biorxiv.org/content/10.1...

@dipcehu.bsky.social
www.biorxiv.org/content/10.1...
Time-dependent and -resolved experiments combined with computation provide a view on molecular dynamics beyond that available from static, ensemble-averaged experiments
Review w @dariagusew.bsky.social & Carl G Henning Hansen
doi.org/10.48550/arX...

Time-dependent and -resolved experiments combined with computation provide a view on molecular dynamics beyond that available from static, ensemble-averaged experiments
Review w @dariagusew.bsky.social & Carl G Henning Hansen
doi.org/10.48550/arX...
The project will focus on molecular modelling of proteins, lipids, and biomolecular condensates at cell membranes.
More details and application form: tinyurl.com/4zm92365
Please feel free to share!
@vetenskapsradet.bsky.social | @mau.se

The project will focus on molecular modelling of proteins, lipids, and biomolecular condensates at cell membranes.
More details and application form: tinyurl.com/4zm92365
Please feel free to share!
@vetenskapsradet.bsky.social | @mau.se
Help publish 3 mature projects, AND develop cool new single molecule fluorescence binding assays!
biophysics
transcription
protein:DNA interactions
my.corehr.com/pls/uoxrecru...

Help publish 3 mature projects, AND develop cool new single molecule fluorescence binding assays!
biophysics
transcription
protein:DNA interactions
my.corehr.com/pls/uoxrecru...
doi.org/10.48550/arX...
doi.org/10.48550/arX...

doi.org/10.48550/arX...
A coarse-grained model for simulations of phosphorylated disordered proteins
(aka parameters for phospho-serine and -threonine for CALVADOS)
is now published in Biophysical Journal
authors.elsevier.com/a/1lTcE1SPTB...
@asrauh.bsky.social @giuliotesei.bsky.social & Gustav Hedemark
A coarse-grained model for simulations of phosphorylated disordered proteins
(aka parameters for phospho-serine and -threonine for CALVADOS)
is now published in Biophysical Journal
authors.elsevier.com/a/1lTcE1SPTB...
@asrauh.bsky.social @giuliotesei.bsky.social & Gustav Hedemark
A coarse-grained model for disordered proteins under crowded conditions
(that is the CALVADOS PEG model) is now published in final form:
dx.doi.org/10.1002/pro....
@asrauh.bsky.social @giuliotesei.bsky.social

A coarse-grained model for disordered proteins under crowded conditions
(that is the CALVADOS PEG model) is now published in final form:
dx.doi.org/10.1002/pro....
@asrauh.bsky.social @giuliotesei.bsky.social
In a fantastic teamwork, @mcagiada.bsky.social and @emilthomasen.bsky.social developed AF2χ to generate conformational ensembles representing side-chain dynamics using AF2 💃
Code: github.com/KULL-Centre/...
Colab: github.com/matteo-cagia...
In a fantastic teamwork, @mcagiada.bsky.social and @emilthomasen.bsky.social developed AF2χ to generate conformational ensembles representing side-chain dynamics using AF2 💃
Code: github.com/KULL-Centre/...
Colab: github.com/matteo-cagia...
Big thanks to @sobuelow.bsky.social, @lindorfflarsen.bsky.social, and the whole team for making this possible.
Thrilled to mark this as my first last-author paper!
We’ve got your back!
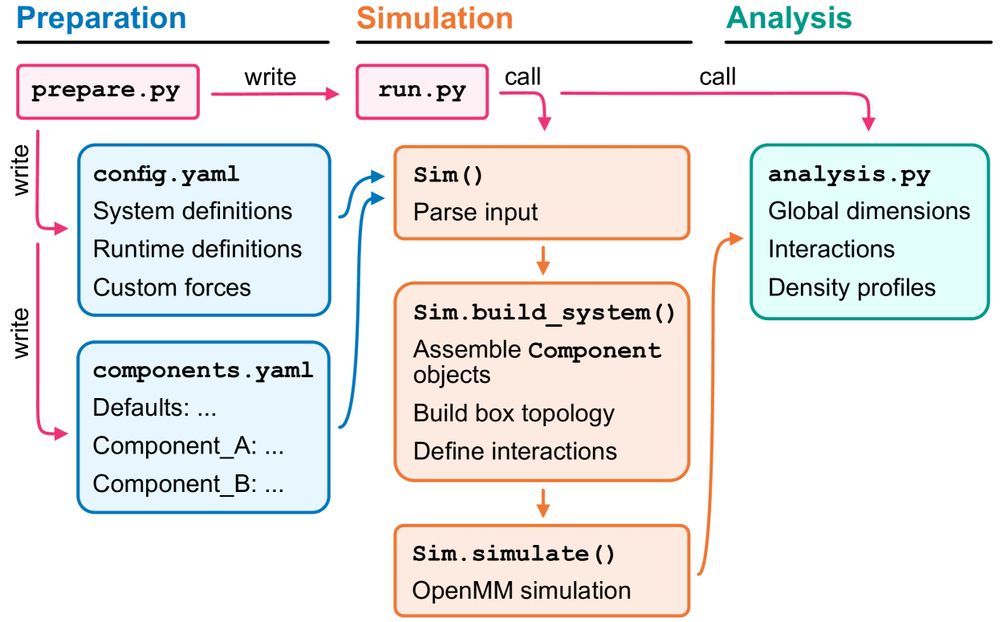
@sobuelow.bsky.social & @giuliotesei.bsky.social—together with the rest of the team—describe our software for simulations using the CALVADOS models incl. recipes for several applications. 1/5
doi.org/10.48550/arX...
Big thanks to @sobuelow.bsky.social, @lindorfflarsen.bsky.social, and the whole team for making this possible.
Thrilled to mark this as my first last-author paper!
We’ve got your back!
@sobuelow.bsky.social & @giuliotesei.bsky.social—together with the rest of the team—describe our software for simulations using the CALVADOS models incl. recipes for several applications. 1/5
doi.org/10.48550/arX...

We’ve got your back!
@sobuelow.bsky.social & @giuliotesei.bsky.social—together with the rest of the team—describe our software for simulations using the CALVADOS models incl. recipes for several applications. 1/5
doi.org/10.48550/arX...
www.pnas.org/doi/10.1073/...
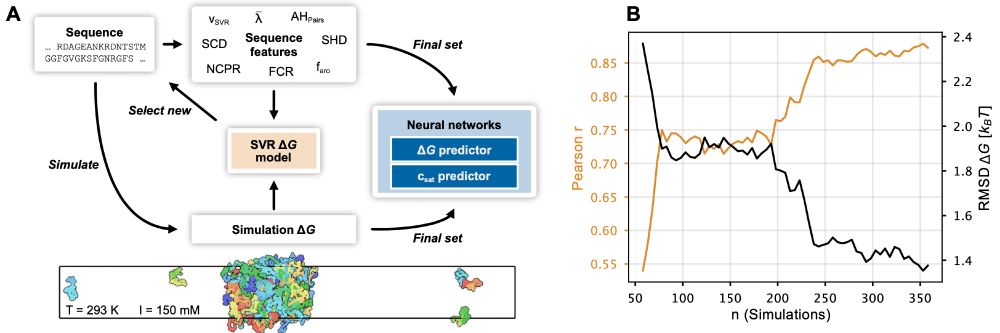
The paper has been substantially updated compared to the preprint including new experimental data and using the neural network to finetune CALVADOS. 1/n
www.pnas.org/doi/10.1073/...
The paper has been substantially updated compared to the preprint including new experimental data and using the neural network to finetune CALVADOS. 1/n
@asrauh.bsky.social @giuliotesei.bsky.social and Gustav Hedemark used a top-down approach in which we targeted experimental data to derive parameters or phosphorylated serine and threonine doi.org/10.1101/2025...

@asrauh.bsky.social @giuliotesei.bsky.social and Gustav Hedemark used a top-down approach in which we targeted experimental data to derive parameters or phosphorylated serine and threonine doi.org/10.1101/2025...
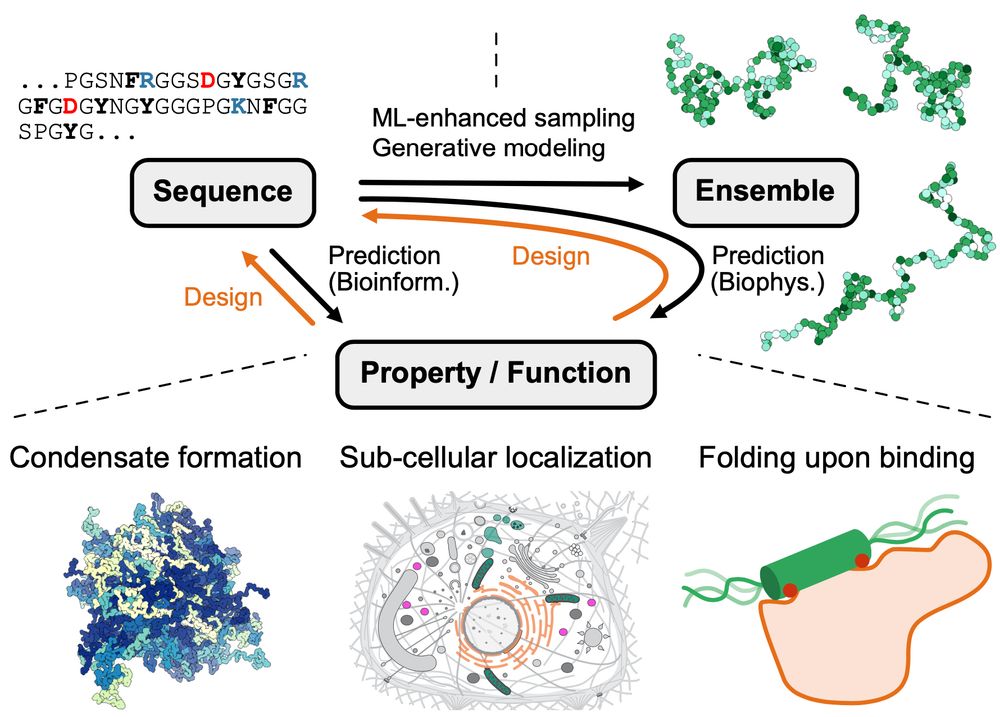
If you are interested in exploring the effects of a crowder on the global dimensions of an IDP or want to explore the phase separation behaviour of a more weakly PS-prone IDP, have a look at our preprint and give it a try.
Work from @asrauh.bsky.social on a simple model for polyethylene glycol to study the effects of crowding on IDPs
If you are interested in exploring the effects of a crowder on the global dimensions of an IDP or want to explore the phase separation behaviour of a more weakly PS-prone IDP, have a look at our preprint and give it a try.
authors.elsevier.com/sd/article/S...
Led by @sobuelow.bsky.social and Giulio Tesei
authors.elsevier.com/sd/article/S...
Led by @sobuelow.bsky.social and Giulio Tesei

