
www.biorxiv.org/content/10.6...
Check out our latest review, out in Advances in Physics X
@juliamaristany.bsky.social @alinaemelianova.bsky.social
@rcollepardo.bsky.social
www.tandfonline.com/doi/epdf/10....

Check out our latest review, out in Advances in Physics X
@juliamaristany.bsky.social @alinaemelianova.bsky.social
@rcollepardo.bsky.social
www.tandfonline.com/doi/epdf/10....
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...

See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
doi.org/10.48550/arX...
doi.org/10.48550/arX...

doi.org/10.48550/arX...
Bad news : current all-atom simulations of phosphorylated IDPs are very probably wrong (and yes, this is clickbaity on purpose 😇)
Good news : we know what to blame for it, and we even have an idea of how to fix it !
Bad news : current all-atom simulations of phosphorylated IDPs are very probably wrong (and yes, this is clickbaity on purpose 😇)
Good news : we know what to blame for it, and we even have an idea of how to fix it !
A coarse-grained model for simulations of phosphorylated disordered proteins
(aka parameters for phospho-serine and -threonine for CALVADOS)
is now published in Biophysical Journal
authors.elsevier.com/a/1lTcE1SPTB...
@asrauh.bsky.social @giuliotesei.bsky.social & Gustav Hedemark
A coarse-grained model for simulations of phosphorylated disordered proteins
(aka parameters for phospho-serine and -threonine for CALVADOS)
is now published in Biophysical Journal
authors.elsevier.com/a/1lTcE1SPTB...
@asrauh.bsky.social @giuliotesei.bsky.social & Gustav Hedemark
A coarse-grained model for disordered proteins under crowded conditions
(that is the CALVADOS PEG model) is now published in final form:
dx.doi.org/10.1002/pro....
@asrauh.bsky.social @giuliotesei.bsky.social

A coarse-grained model for disordered proteins under crowded conditions
(that is the CALVADOS PEG model) is now published in final form:
dx.doi.org/10.1002/pro....
@asrauh.bsky.social @giuliotesei.bsky.social
In a fantastic teamwork, @mcagiada.bsky.social and @emilthomasen.bsky.social developed AF2χ to generate conformational ensembles representing side-chain dynamics using AF2 💃
Code: github.com/KULL-Centre/...
Colab: github.com/matteo-cagia...
In a fantastic teamwork, @mcagiada.bsky.social and @emilthomasen.bsky.social developed AF2χ to generate conformational ensembles representing side-chain dynamics using AF2 💃
Code: github.com/KULL-Centre/...
Colab: github.com/matteo-cagia...
Give it a read: doi.org/10.48550/arX...
Give it a try: github.com/KULL-Centre/...
Give it a read: doi.org/10.48550/arX...
Give it a try: github.com/KULL-Centre/...
Check out the published version (including added exp. data from @tanjamittag.bsky.social) and feel free to try out our webserver.
www.pnas.org/doi/10.1073/...
The paper has been substantially updated compared to the preprint including new experimental data and using the neural network to finetune CALVADOS. 1/n
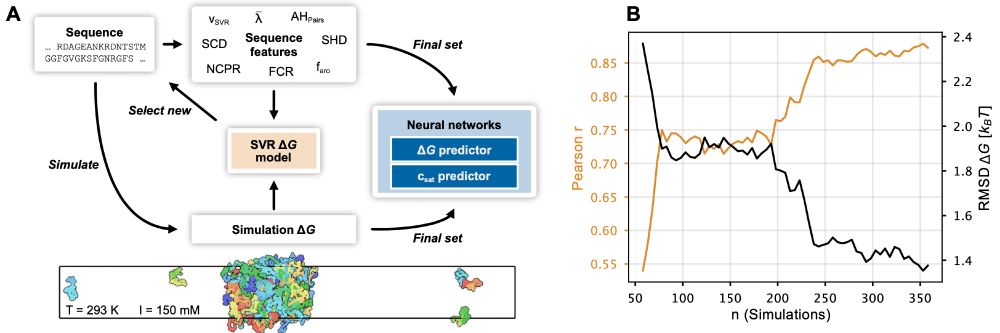
Check out the published version (including added exp. data from @tanjamittag.bsky.social) and feel free to try out our webserver.
If you want to explore the changes in global dimensions of a disordered protein upon phosphorylation: give it a read and a try!
Big thank you to @giuliotesei.bsky.social, @lindorfflarsen.bsky.social and Gustav S. Hedemark
@asrauh.bsky.social @giuliotesei.bsky.social and Gustav Hedemark used a top-down approach in which we targeted experimental data to derive parameters or phosphorylated serine and threonine doi.org/10.1101/2025...
If you want to explore the changes in global dimensions of a disordered protein upon phosphorylation: give it a read and a try!
Big thank you to @giuliotesei.bsky.social, @lindorfflarsen.bsky.social and Gustav S. Hedemark
If you are interested in exploring the effects of a crowder on the global dimensions of an IDP or want to explore the phase separation behaviour of a more weakly PS-prone IDP, have a look at our preprint and give it a try.
Work from @asrauh.bsky.social on a simple model for polyethylene glycol to study the effects of crowding on IDPs
If you are interested in exploring the effects of a crowder on the global dimensions of an IDP or want to explore the phase separation behaviour of a more weakly PS-prone IDP, have a look at our preprint and give it a try.

Paper led by Jacob Aunstrup from Alex Büll’s lab with MD simulations by Abigail Barclay, and key contributions from several others. We combined measurements of Φ-values with MD simulations to study the transition state for amyloid fibril growth
doi.org/10.1038/s415...

Paper led by Jacob Aunstrup from Alex Büll’s lab with MD simulations by Abigail Barclay, and key contributions from several others. We combined measurements of Φ-values with MD simulations to study the transition state for amyloid fibril growth
doi.org/10.1038/s415...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Link to paper: www.cell.com/structure/fu...
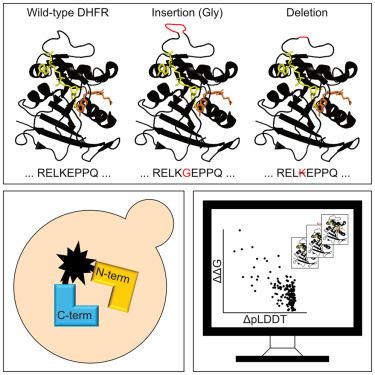
Link to paper: www.cell.com/structure/fu...
Ikki Yasuda, Sören von Bülow & Giulio Tesei have parameterized a simple model for disordered RNA. Despite it's simplicity (no sequence, no base pairing) we find that it captures several phenomena that depend on the charge, stickiness and polymer properties of RNA 🧬🧶🧪

Ikki Yasuda, Sören von Bülow & Giulio Tesei have parameterized a simple model for disordered RNA. Despite it's simplicity (no sequence, no base pairing) we find that it captures several phenomena that depend on the charge, stickiness and polymer properties of RNA 🧬🧶🧪
Details: phdcourses.ku.dk/DetailKursus...

Details: phdcourses.ku.dk/DetailKursus...
Identifying Sequence Effects on Chain Dimensions of Disordered Proteins by Integrating Experiments and Simulations.
pubs.acs.org/doi/10.1021/...
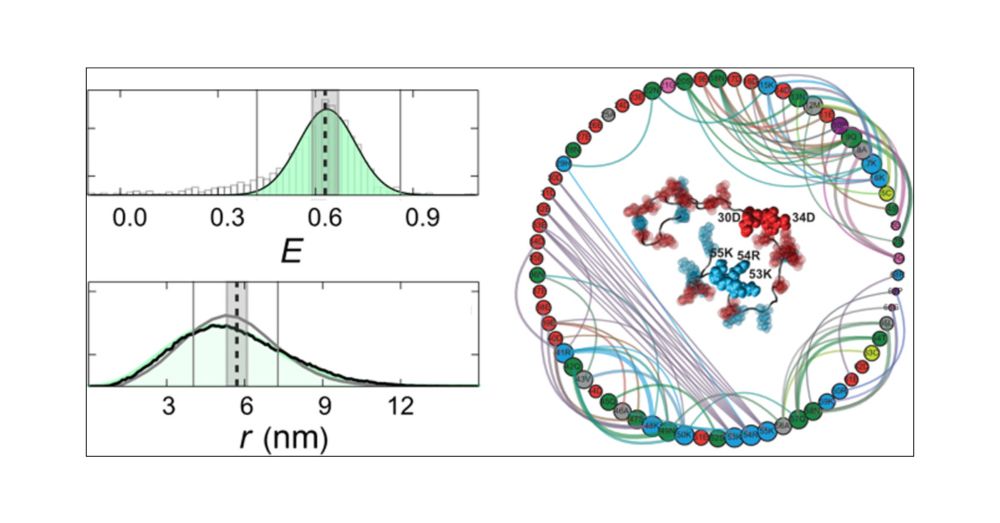
Identifying Sequence Effects on Chain Dimensions of Disordered Proteins by Integrating Experiments and Simulations.
pubs.acs.org/doi/10.1021/...


