
See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...

See preprint for:
— Ensembles of >12000 full-length human proteins
— Analysis of IDRs in >1500 TFs
📜 doi.org/10.1101/2025...
💾 github.com/KULL-Centre/...
A coarse-grained model for disordered proteins under crowded conditions
(that is the CALVADOS PEG model) is now published in final form:
dx.doi.org/10.1002/pro....
@asrauh.bsky.social @giuliotesei.bsky.social

A coarse-grained model for disordered proteins under crowded conditions
(that is the CALVADOS PEG model) is now published in final form:
dx.doi.org/10.1002/pro....
@asrauh.bsky.social @giuliotesei.bsky.social
We are actively working improving/updating various aspects of FINCHES; don't hesitate to reach out if you run into issues, have questions.
www.science.org/doi/10.1126/...
We are actively working improving/updating various aspects of FINCHES; don't hesitate to reach out if you run into issues, have questions.
www.science.org/doi/10.1126/...
We’ve got your back!
@sobuelow.bsky.social & @giuliotesei.bsky.social—together with the rest of the team—describe our software for simulations using the CALVADOS models incl. recipes for several applications. 1/5
doi.org/10.48550/arX...
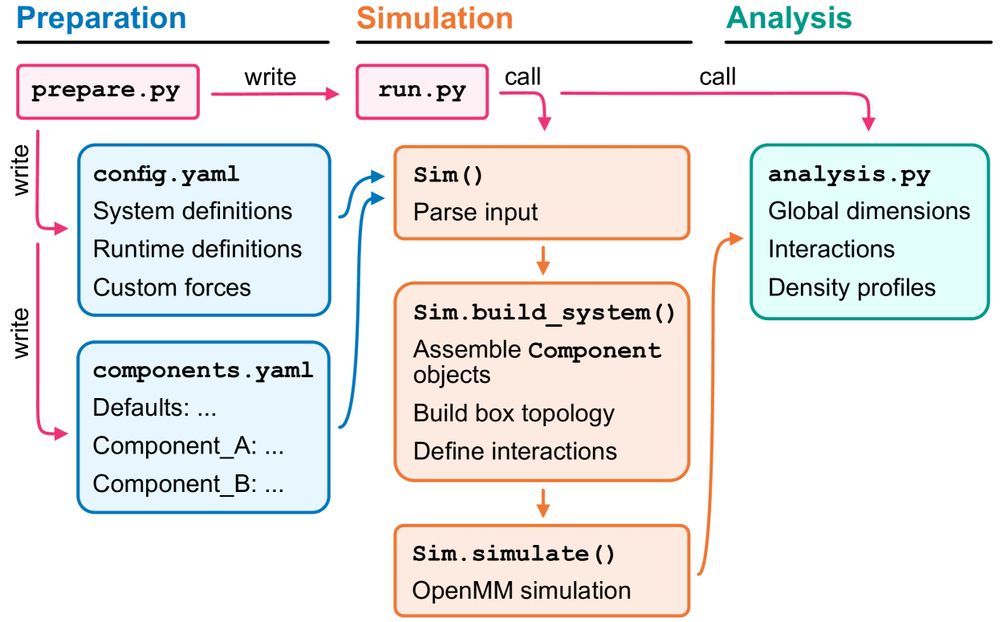
We’ve got your back!
@sobuelow.bsky.social & @giuliotesei.bsky.social—together with the rest of the team—describe our software for simulations using the CALVADOS models incl. recipes for several applications. 1/5
doi.org/10.48550/arX...
Max Planck Research Group Leader (W2) in Molecular Design
...and make your own research dreams happen on de novo design, generative models, proteins, materials ...
tinyurl.com/r2xjxnuk
@mpip-mainz.mpg.de

Max Planck Research Group Leader (W2) in Molecular Design
...and make your own research dreams happen on de novo design, generative models, proteins, materials ...
tinyurl.com/r2xjxnuk
@mpip-mainz.mpg.de
Here @tkschulze.bsky.social devised a strategy to take this into account while training models
www.biorxiv.org/content/10.1...

Here @tkschulze.bsky.social devised a strategy to take this into account while training models
www.biorxiv.org/content/10.1...
Check out the published version (including added exp. data from @tanjamittag.bsky.social) and feel free to try out our webserver.
www.pnas.org/doi/10.1073/...
The paper has been substantially updated compared to the preprint including new experimental data and using the neural network to finetune CALVADOS. 1/n
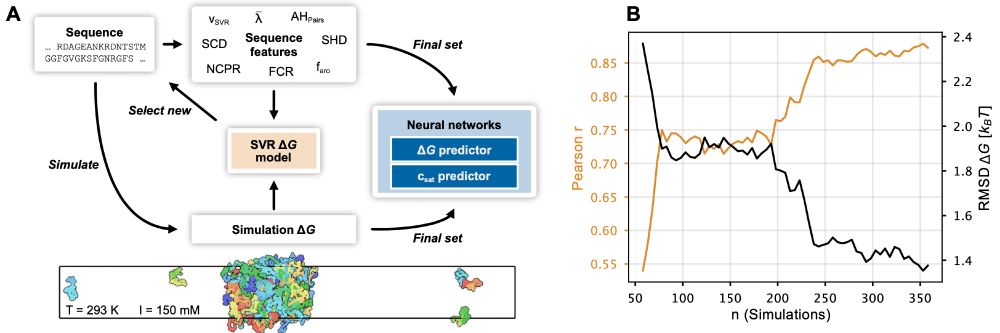
Check out the published version (including added exp. data from @tanjamittag.bsky.social) and feel free to try out our webserver.
authors.elsevier.com/sd/article/S...
Led by @sobuelow.bsky.social and Giulio Tesei
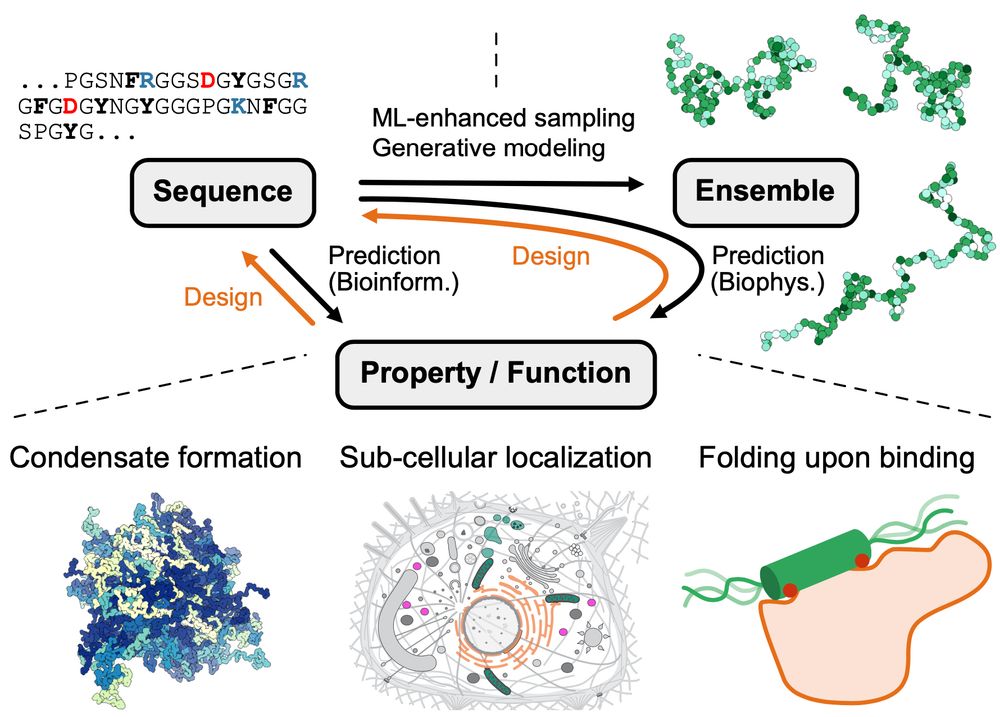
authors.elsevier.com/sd/article/S...
Led by @sobuelow.bsky.social and Giulio Tesei
Work from @asrauh.bsky.social on a simple model for polyethylene glycol to study the effects of crowding on IDPs

Work from @asrauh.bsky.social on a simple model for polyethylene glycol to study the effects of crowding on IDPs
doi.org/10.1021/acs....
This is a simple model for flexible RNA that complements and works with the CALVADOS protein model. Work led by Ikki Yasuda who visited us from Keio University.
Try it yourself using our latest code for CALVADOS
github.com/KULL-Centre/...
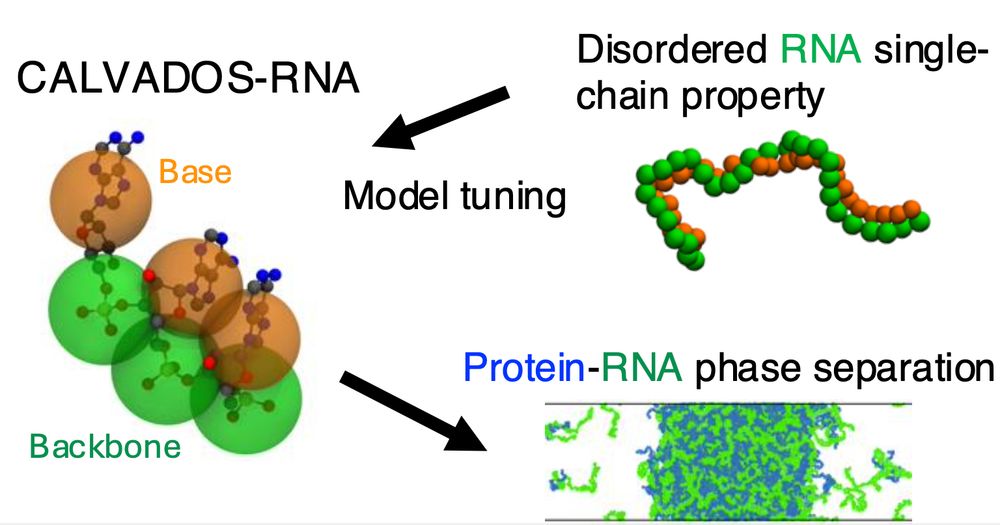
doi.org/10.1021/acs....
This is a simple model for flexible RNA that complements and works with the CALVADOS protein model. Work led by Ikki Yasuda who visited us from Keio University.
Try it yourself using our latest code for CALVADOS
github.com/KULL-Centre/...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Ikki Yasuda, Sören von Bülow & Giulio Tesei have parameterized a simple model for disordered RNA. Despite it's simplicity (no sequence, no base pairing) we find that it captures several phenomena that depend on the charge, stickiness and polymer properties of RNA 🧬🧶🧪

Ikki Yasuda, Sören von Bülow & Giulio Tesei have parameterized a simple model for disordered RNA. Despite it's simplicity (no sequence, no base pairing) we find that it captures several phenomena that depend on the charge, stickiness and polymer properties of RNA 🧬🧶🧪
go.bsky.app/J23B51L
go.bsky.app/J23B51L
We find that much of the variation can be explained and predicted by a burial-dependent substitution matrix
Lots more goodies in the paper
doi.org/10.1101/2024...

We find that much of the variation can be explained and predicted by a burial-dependent substitution matrix
Lots more goodies in the paper
doi.org/10.1101/2024...
We combined active learning and coarse-grained simulations to develop a machine learning model for quantitative predictions of IDR phase separation 🧬🧶
doi.org/10.1101/2024...

We combined active learning and coarse-grained simulations to develop a machine learning model for quantitative predictions of IDR phase separation 🧬🧶
doi.org/10.1101/2024...
We show that a Calpha representation of the folded domains can give rise to too compact conformations of multi-domain proteins (MDPs), and that a centre-of-mass representation in the folded domains improves agreement with experiments. 🧬🧶

We show that a Calpha representation of the folded domains can give rise to too compact conformations of multi-domain proteins (MDPs), and that a centre-of-mass representation in the folded domains improves agreement with experiments. 🧬🧶

