Rémi Trimbour - MD-PhD student
@remitrimbour.bsky.social
MD-PhD student, computational biology / single-cell omics
Currently at
@cantinilab.bsky.social - Institut Pasteur & @saezlab.bsky.social - Heidelberg Uni
Tools: https://www.github.com/r-trimbour
Publications: https://tinyurl.com/TrimbourRemi
Currently at
@cantinilab.bsky.social - Institut Pasteur & @saezlab.bsky.social - Heidelberg Uni
Tools: https://www.github.com/r-trimbour
Publications: https://tinyurl.com/TrimbourRemi
I'm presenting #CIRCE, a Python package to infer co-accessible DNA region networks 🧬
Based on #Cicero 's algorithm (Pliner et al.), it runs ~150x faster, processing an atlas of 700k cells in less than 40 min! ⛷️
Short paper: doi.org/10.1101/2025...
Code: github.com/cantinilab/CIRCE
1/5 ⬇️
Based on #Cicero 's algorithm (Pliner et al.), it runs ~150x faster, processing an atlas of 700k cells in less than 40 min! ⛷️
Short paper: doi.org/10.1101/2025...
Code: github.com/cantinilab/CIRCE
1/5 ⬇️

September 30, 2025 at 12:23 PM
I'm presenting #CIRCE, a Python package to infer co-accessible DNA region networks 🧬
Based on #Cicero 's algorithm (Pliner et al.), it runs ~150x faster, processing an atlas of 700k cells in less than 40 min! ⛷️
Short paper: doi.org/10.1101/2025...
Code: github.com/cantinilab/CIRCE
1/5 ⬇️
Based on #Cicero 's algorithm (Pliner et al.), it runs ~150x faster, processing an atlas of 700k cells in less than 40 min! ⛷️
Short paper: doi.org/10.1101/2025...
Code: github.com/cantinilab/CIRCE
1/5 ⬇️
Reposted by Rémi Trimbour - MD-PhD student
I'm curious if other mid career PIs have considered completely abandoning the trad publication model. We've been making gradual moves towards that end goal. But it's been difficult to completely jump ship for one main reason for me at least. 1/
September 16, 2025 at 5:43 AM
I'm curious if other mid career PIs have considered completely abandoning the trad publication model. We've been making gradual moves towards that end goal. But it's been difficult to completely jump ship for one main reason for me at least. 1/
Reposted by Rémi Trimbour - MD-PhD student
On Tuesday at 2025-02-04 18:00 CET, @jkobject will talk about scPrint, a transformer model that infers gene networks from scRNA-seq data, at our 2nd community meeting of 2025! For more information, check out the GitHub: https://buff.ly/40RJ3gT & pre-print https://buff.ly/4hwK4Av

GitHub - cantinilab/scPRINT: single cell foundation model for Gene network inference and more
single cell foundation model for Gene network inference and more - cantinilab/scPRINT
buff.ly
January 31, 2025 at 6:02 PM
On Tuesday at 2025-02-04 18:00 CET, @jkobject will talk about scPrint, a transformer model that infers gene networks from scRNA-seq data, at our 2nd community meeting of 2025! For more information, check out the GitHub: https://buff.ly/40RJ3gT & pre-print https://buff.ly/4hwK4Av
Reposted by Rémi Trimbour - MD-PhD student
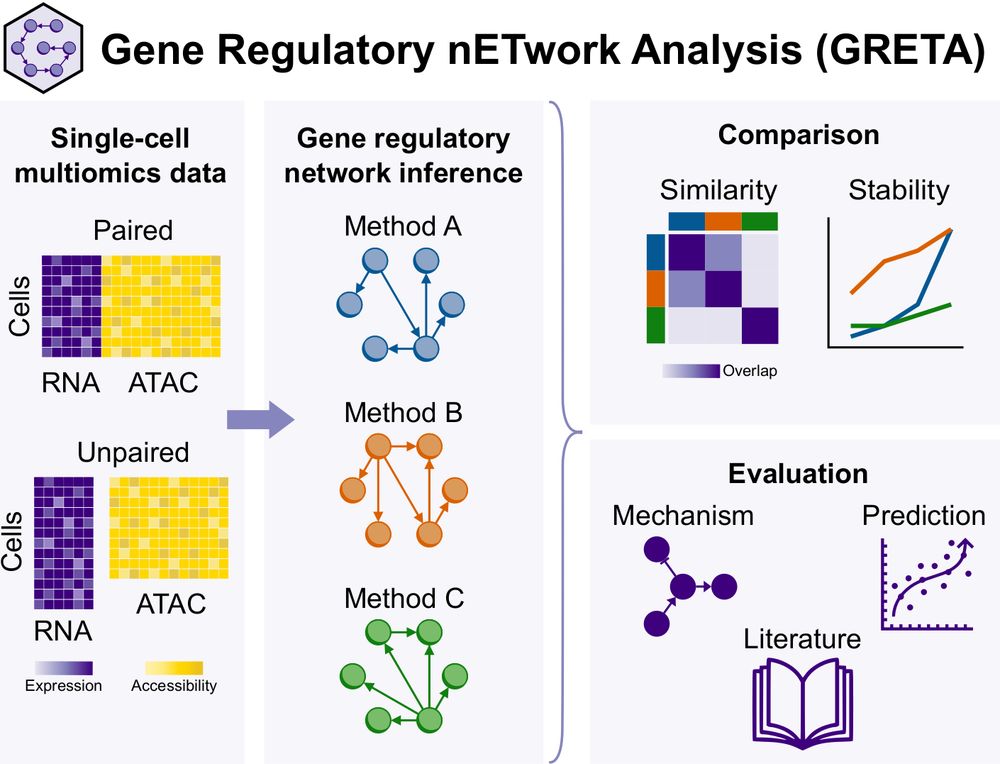
We present Gene Regulatory nETwork Analsyis (GRETA), a framework to infer, compare and evaluate gene regulatory networks #GRNs. With it, we have benchmarked multimodal and unimodal GRN inference methods. Check the results here 👇
Paper: doi.org/10.1101/2024.12.20.629764
Code: github.com/saezlab/greta
Paper: doi.org/10.1101/2024.12.20.629764
Code: github.com/saezlab/greta
December 23, 2024 at 8:43 AM
We present Gene Regulatory nETwork Analsyis (GRETA), a framework to infer, compare and evaluate gene regulatory networks #GRNs. With it, we have benchmarked multimodal and unimodal GRN inference methods. Check the results here 👇
Paper: doi.org/10.1101/2024.12.20.629764
Code: github.com/saezlab/greta
Paper: doi.org/10.1101/2024.12.20.629764
Code: github.com/saezlab/greta
Reposted by Rémi Trimbour - MD-PhD student
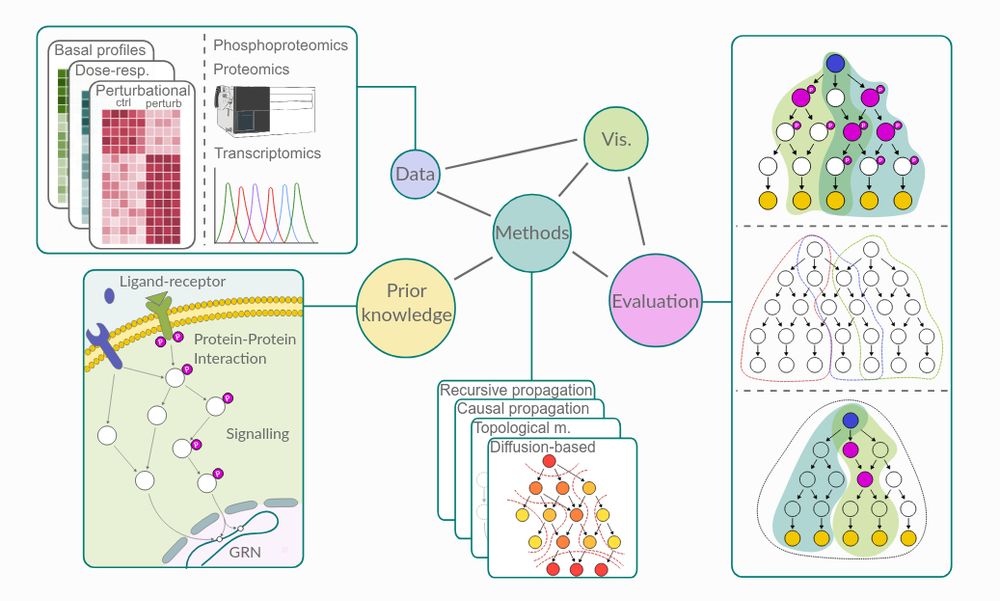
We present NetworkCommons, a unified platform 🪐 for network biology, providing access to omics data, knowledge, and contextualization methods, all with a consistent API 👇🧵
Paper: doi.org/10.1101/2024...
Docs: networkcommons.readthedocs.io
Paper: doi.org/10.1101/2024...
Docs: networkcommons.readthedocs.io
November 26, 2024 at 9:44 AM
We present NetworkCommons, a unified platform 🪐 for network biology, providing access to omics data, knowledge, and contextualization methods, all with a consistent API 👇🧵
Paper: doi.org/10.1101/2024...
Docs: networkcommons.readthedocs.io
Paper: doi.org/10.1101/2024...
Docs: networkcommons.readthedocs.io
Reposted by Rémi Trimbour - MD-PhD student
Hey, this place is getting fun! Here is a new version of our latest preprint on oscillatory dynamics of mRNA metabolism and chromatin accessibility during the cell cycle: www.biorxiv.org/content/10.1...

Single-cell multiomics reveals the oscillatory dynamics of mRNA metabolism and chromatin accessibility during the cell cycle
The cell cycle is a tightly regulated process that requires precise temporal expression of hundreds of cell cycledependent genes. However, the genome-wide dynamics of mRNA metabolism throughout the ce...
www.biorxiv.org
November 22, 2024 at 11:04 AM
Hey, this place is getting fun! Here is a new version of our latest preprint on oscillatory dynamics of mRNA metabolism and chromatin accessibility during the cell cycle: www.biorxiv.org/content/10.1...
Reposted by Rémi Trimbour - MD-PhD student
What are the key disrupted multicellular processes in heart failure? In our new work we combine 23 years of molecular data with recent single-cell atlases to draw a cross-study patient map
doi.org/10.1101/2024...
doi.org/10.1101/2024...

November 11, 2024 at 9:07 AM
What are the key disrupted multicellular processes in heart failure? In our new work we combine 23 years of molecular data with recent single-cell atlases to draw a cross-study patient map
doi.org/10.1101/2024...
doi.org/10.1101/2024...

