1/6
By A. Oravecz and our collaborators @molinalab.bsky.social

By A. Oravecz and our collaborators @molinalab.bsky.social


doi.org/10.1093/nar/...
Find out how we can reconstruct enhancer activity in vivo in the Drosophila embryo using scRNAseq data and Optimal Transport.
doi.org/10.1093/nar/...
Find out how we can reconstruct enhancer activity in vivo in the Drosophila embryo using scRNAseq data and Optimal Transport.

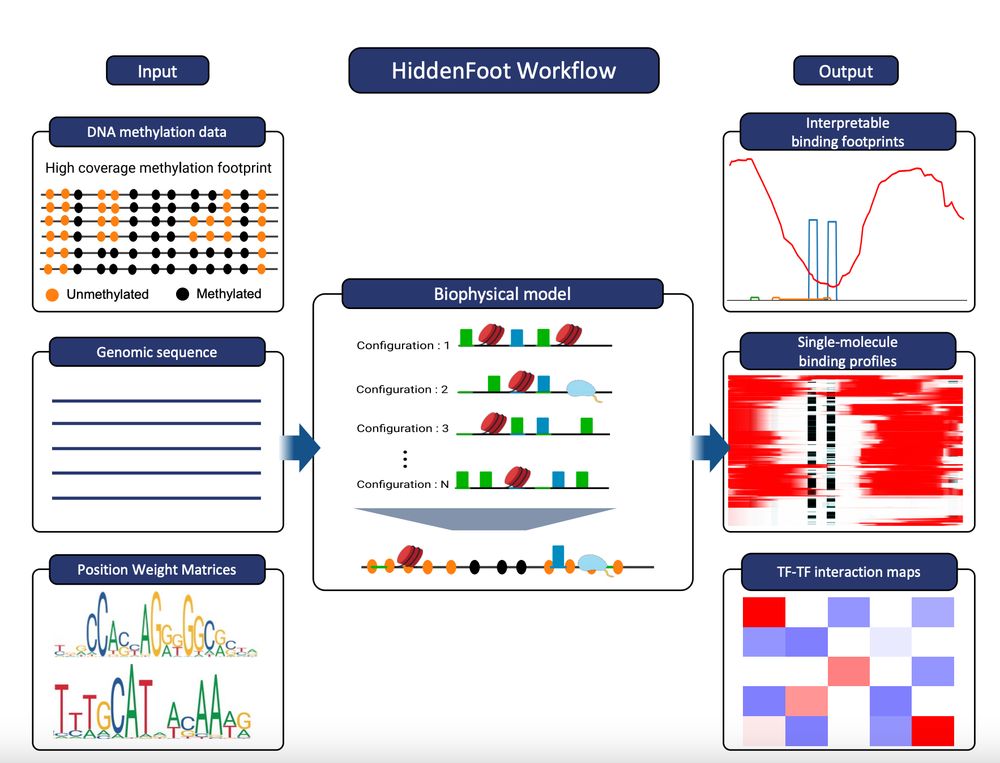
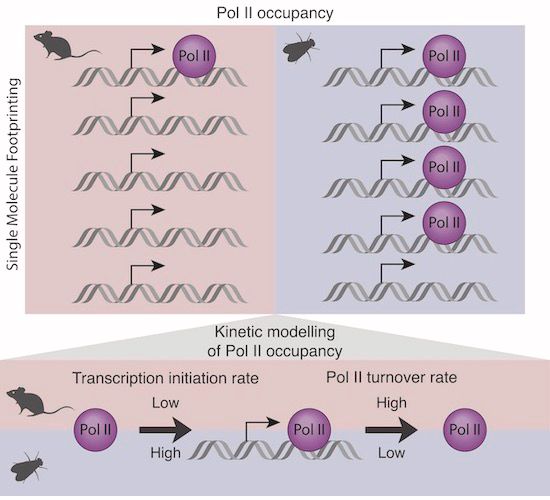
➡️ 'Decoding Gene Regulation at the Single-Molecule Level through Biophysical Modeling of DNA Footprinting Data'
🎙️ Nacho Molina – Institute of Genetics and Molecular and Cellular Biology
@molinalab.bsky.social @igbmc.bsky.social @embl.org



➡️ 'Decoding Gene Regulation at the Single-Molecule Level through Biophysical Modeling of DNA Footprinting Data'
🎙️ Nacho Molina – Institute of Genetics and Molecular and Cellular Biology
@molinalab.bsky.social @igbmc.bsky.social @embl.org
𝐐𝐮𝐚𝐧𝐭𝐢𝐟𝐢𝐜𝐚𝐭𝐢𝐨𝐧 𝐨𝐟 𝐠𝐞𝐧𝐞 𝐞𝐱𝐩𝐫𝐞𝐬𝐬𝐢𝐨𝐧 𝐜𝐨𝐧𝐭𝐫𝐨𝐥 𝐢𝐧 𝐚 𝐦𝐚𝐦𝐦𝐚𝐥𝐢𝐚𝐧 𝐭𝐢𝐬𝐬𝐮𝐞 𝐚𝐭 𝐬𝐢𝐧𝐠𝐥𝐞 𝐜𝐞𝐥𝐥 𝐫𝐞𝐬𝐨𝐥𝐮𝐭𝐢𝐨𝐧
youtu.be/adkY6txDyqs?...
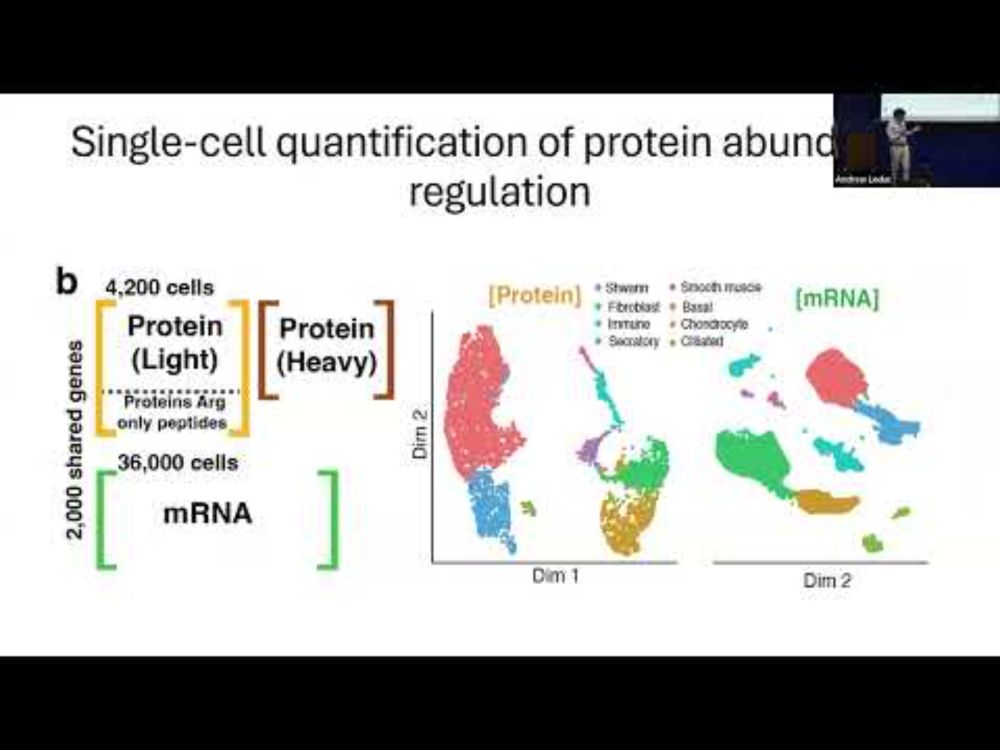
𝐐𝐮𝐚𝐧𝐭𝐢𝐟𝐢𝐜𝐚𝐭𝐢𝐨𝐧 𝐨𝐟 𝐠𝐞𝐧𝐞 𝐞𝐱𝐩𝐫𝐞𝐬𝐬𝐢𝐨𝐧 𝐜𝐨𝐧𝐭𝐫𝐨𝐥 𝐢𝐧 𝐚 𝐦𝐚𝐦𝐦𝐚𝐥𝐢𝐚𝐧 𝐭𝐢𝐬𝐬𝐮𝐞 𝐚𝐭 𝐬𝐢𝐧𝐠𝐥𝐞 𝐜𝐞𝐥𝐥 𝐫𝐞𝐬𝐨𝐥𝐮𝐭𝐢𝐨𝐧
youtu.be/adkY6txDyqs?...
Whether you are new or an established researcher in the Basel-Freiburg-Strasbourg region, this is your meeting when working in transcription, chromatin & gene regulation!
✍️ www.ie-freiburg.mpg.de/gene-regulat...
Whether you are new or an established researcher in the Basel-Freiburg-Strasbourg region, this is your meeting when working in transcription, chromatin & gene regulation!
✍️ www.ie-freiburg.mpg.de/gene-regulat...
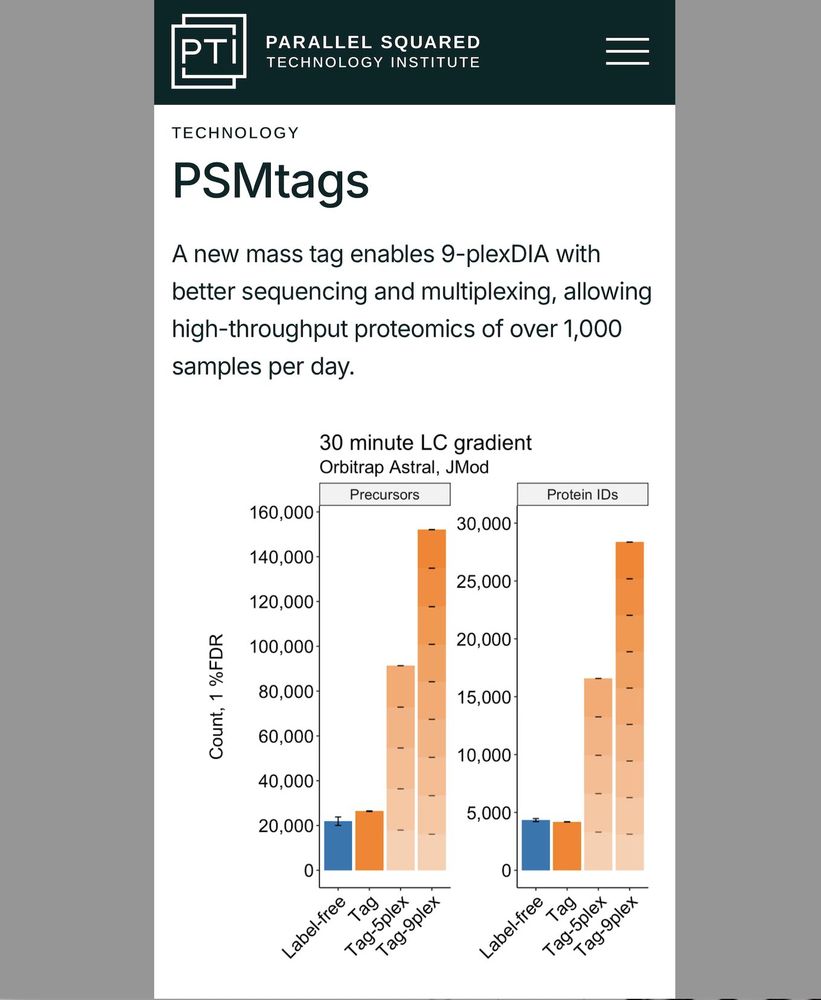
🟧 Multiplexed single-cell proteomics affords higher throughput (orange bars).
⬛️ Proteome depth & quantitative accuracy are comparable.
We aim to make the 🟧 bars taller.
www.parallelsq.org/psmtags
www.biorxiv.org/content/10.1...
🟧 Multiplexed single-cell proteomics affords higher throughput (orange bars).
⬛️ Proteome depth & quantitative accuracy are comparable.
We aim to make the 🟧 bars taller.
www.parallelsq.org/psmtags
www.biorxiv.org/content/10.1...
S02E05 Apocalipsis en la historia (parte I)
Spotify
open.spotify.com/episode/7CJs...
YouTube
youtu.be/fTa3DROe1vE
iVoox
go.ivoox.com/rf/148751814
Y Apple Podcasts o cualquier app que te guste. ¡Amárrate al asiento, que hoy el programa va lleno de desastres! #podcast
🎫 Register by 3 Jun 👉🏼 http://s.embl.org/grg25-01-bl
🧬RNA processing
🧬Translation
🧬Transcription & chromatin regulation
🧬Method development
🧬Theory

🎫 Register by 3 Jun 👉🏼 http://s.embl.org/grg25-01-bl
🧬RNA processing
🧬Translation
🧬Transcription & chromatin regulation
🧬Method development
🧬Theory

🎫 Register by 3 Jun 👉🏼 http://s.embl.org/grg25-01-bl
🧬RNA processing
🧬Translation
🧬Transcription & chromatin regulation
🧬Method development
🧬Theory
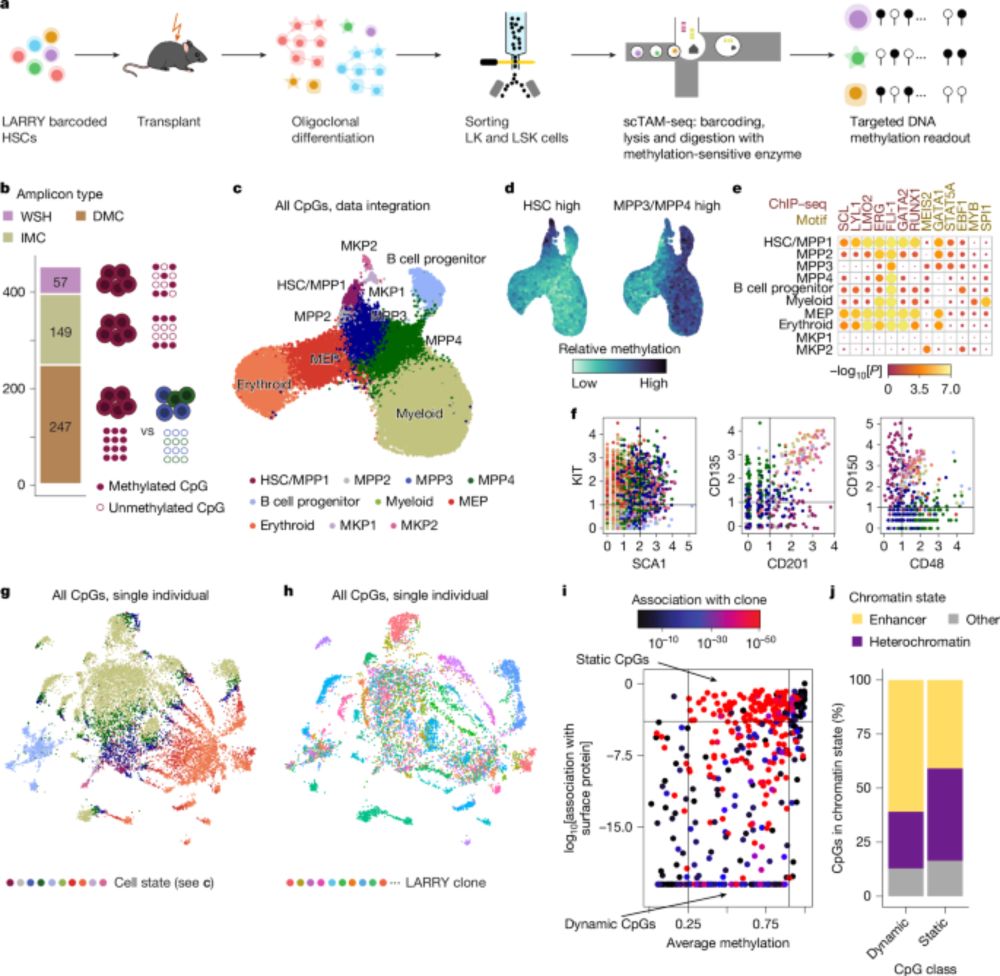
🧬 Single cell methylome encodes cell state & clonal identity
🔨 EPI-Clone reads out both (+mutations, +RNA) at scale
🩸 Clonal expansions of HSCs are universal from age 50, not driven by CH mutations
doi.org/10.1038/s415...
🧵
🧬 Single cell methylome encodes cell state & clonal identity
🔨 EPI-Clone reads out both (+mutations, +RNA) at scale
🩸 Clonal expansions of HSCs are universal from age 50, not driven by CH mutations
doi.org/10.1038/s415...
🧵
1/6

1/6

1/6

Many congratulations 👏🏻 🎉 to all those involved!
Hey #ChromatinSky people where are you at 🤩?
1/6

Many congratulations 👏🏻 🎉 to all those involved!
Hey #ChromatinSky people where are you at 🤩?
1/6

1/6

I'm so excited about this!
www.biorxiv.org/content/10.1...

I'm so excited about this!
www.biorxiv.org/content/10.1...
Details here: www.fondation-arc.org/projets/recr...
Interested? Contact us!
Details here: www.fondation-arc.org/projets/recr...
Interested? Contact us!
Very happy to see this come to fruition- Look forward to feedback from colleagues !
More 👉 www.cell.com/cell/fulltex...

Very happy to see this come to fruition- Look forward to feedback from colleagues !
More 👉 www.cell.com/cell/fulltex...
www.biorxiv.org/cgi/content/...

www.biorxiv.org/cgi/content/...

