Nacho Molina
@molinalab.bsky.social
Group leader of the Stochastic Systems Biology Lab at IGBMC - CNRS - University of Strasbourg. Models of gene regulation based on biophysics-informed deep learning: https://www.igbmc.fr/molina
Finally, if you've made it this far, you might be interested in testing the code. Feedback is very welcome! github.com/MolinaLab-IG...
6/6
6/6

May 19, 2025 at 7:29 PM
Finally, if you've made it this far, you might be interested in testing the code. Feedback is very welcome! github.com/MolinaLab-IG...
6/6
6/6
HiddenFoot also works with Fiber-seq data! It reveals chromatin structure and nucleosome occupancy heterogeneity at single-molecule resolution driven by TF binding.
5/6
5/6

May 19, 2025 at 7:24 PM
HiddenFoot also works with Fiber-seq data! It reveals chromatin structure and nucleosome occupancy heterogeneity at single-molecule resolution driven by TF binding.
5/6
5/6
HiddenFoot resolves Pol II and nucleosome occupancy at the HIV-1 promoter, molecule by molecule. Under transcriptional inhibition (TLD), Pol II footprints vanish, while nucleosome occupancy at the +1 and −1 positions increases.
4/6
4/6

May 19, 2025 at 7:19 PM
HiddenFoot resolves Pol II and nucleosome occupancy at the HIV-1 promoter, molecule by molecule. Under transcriptional inhibition (TLD), Pol II footprints vanish, while nucleosome occupancy at the +1 and −1 positions increases.
4/6
4/6
HiddenFoot infers pairwise interaction energies from single-molecule data and compares them to simulated equilibrium profiles to distinguish true TF–TF cooperativity from nucleosome-mediated co-binding.
3/6
3/6

May 19, 2025 at 7:19 PM
HiddenFoot infers pairwise interaction energies from single-molecule data and compares them to simulated equilibrium profiles to distinguish true TF–TF cooperativity from nucleosome-mediated co-binding.
3/6
3/6
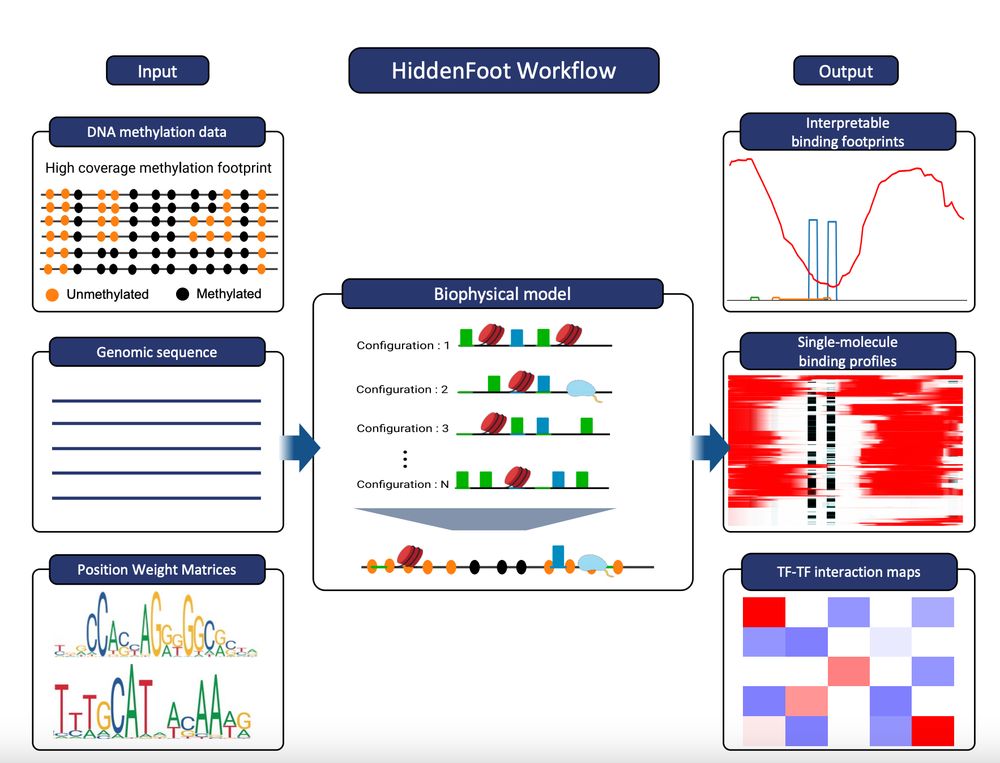
HiddenFoot is a thermodynamics-based model that integrates known TF PWMs and nucleosome occupancy to efficiently evaluate all possible non-overlapping binding configurations. It fits model parameters using both stochastic gradient descent and MCMC.
2/6
2/6

May 19, 2025 at 7:07 PM
HiddenFoot is a thermodynamics-based model that integrates known TF PWMs and nucleosome occupancy to efficiently evaluate all possible non-overlapping binding configurations. It fits model parameters using both stochastic gradient descent and MCMC.
2/6
2/6
Time for a short thread! We developed HiddenFoot, a biophysics-inspired approach to decode single-molecule footprinting data and infer TF, nucleosome, and RNA Pol II binding profiles on individual DNA molecules. One molecule at a time! www.biorxiv.org/content/10.1...
1/6
1/6
May 19, 2025 at 6:55 PM
Time for a short thread! We developed HiddenFoot, a biophysics-inspired approach to decode single-molecule footprinting data and infer TF, nucleosome, and RNA Pol II binding profiles on individual DNA molecules. One molecule at a time! www.biorxiv.org/content/10.1...
1/6
1/6
🚨 New preprint out! Do you think Single Molecule Footprinting and Fiber-seq are super cool but aren't sure how to unlock their full potential? HiddenFoot can help you: www.biorxiv.org/content/10.1...

May 17, 2025 at 5:00 PM
🚨 New preprint out! Do you think Single Molecule Footprinting and Fiber-seq are super cool but aren't sure how to unlock their full potential? HiddenFoot can help you: www.biorxiv.org/content/10.1...
Hey! Check out our latest publication. A great collaboration with @longchrom.bsky.social and a nice example of how a negative result, the absence of a response to a transcriptional perturbation, can actually reveal an intriguing gene-specific buffering mechanism.
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...

March 31, 2025 at 7:30 AM
Hey! Check out our latest publication. A great collaboration with @longchrom.bsky.social and a nice example of how a negative result, the absence of a response to a transcriptional perturbation, can actually reveal an intriguing gene-specific buffering mechanism.
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
3) Waves of RNA degradation rates and top predicted RBPs: Similarly, Zfp36l1, Tia1, Hnrnpl, Nudt21, Srsf1, and Fus have been linked to regulation of RNA stability or/and cell cycle... So, perhaps our predictions are not completely random! Happy to hear your feedback.

November 26, 2024 at 9:03 PM
3) Waves of RNA degradation rates and top predicted RBPs: Similarly, Zfp36l1, Tia1, Hnrnpl, Nudt21, Srsf1, and Fus have been linked to regulation of RNA stability or/and cell cycle... So, perhaps our predictions are not completely random! Happy to hear your feedback.
2) Waves of RNA export rates and top predicted RBPs: It seems that Nxf1, Hnrnpk, Taf15, Rbm10, Strap, and Ep300 may have a role in RNA export or/and cell cycle regulation.

November 26, 2024 at 8:59 PM
2) Waves of RNA export rates and top predicted RBPs: It seems that Nxf1, Hnrnpk, Taf15, Rbm10, Strap, and Ep300 may have a role in RNA export or/and cell cycle regulation.
1) Waves of transcription rates and top predicted TFs: Previous evidence shows that E2f4, Hmga1, Mybl1, and Hes1 are linked to cell cycle control and proliferation.

November 26, 2024 at 8:58 PM
1) Waves of transcription rates and top predicted TFs: Previous evidence shows that E2f4, Hmga1, Mybl1, and Hes1 are linked to cell cycle control and proliferation.
My favourite place for conferences!

November 19, 2024 at 12:28 PM
My favourite place for conferences!
Welcome! Avez-vous un compte en anglais ?

January 18, 2024 at 9:54 AM
Welcome! Avez-vous un compte en anglais ?
10X multiome allowed us to assess how chromatin accessibility changes throughout the cell cycle. Interestingly, dynamics within TF footprints reveal key cell cycle regulators.

January 17, 2024 at 10:31 AM
10X multiome allowed us to assess how chromatin accessibility changes throughout the cell cycle. Interestingly, dynamics within TF footprints reveal key cell cycle regulators.
Extending our biophysical model to single-nucleus transcriptomes shows clear waves of nuclear mRNA export!

January 17, 2024 at 10:30 AM
Extending our biophysical model to single-nucleus transcriptomes shows clear waves of nuclear mRNA export!
Genome-wide, we've identified distinct waves of transcription and degradation during the cell cycle. Notably, postranscriptional regulation emerges as a key player in shaping mRNA accumulation.

January 17, 2024 at 10:30 AM
Genome-wide, we've identified distinct waves of transcription and degradation during the cell cycle. Notably, postranscriptional regulation emerges as a key player in shaping mRNA accumulation.
Fitting a biophysical model to unspliced and spliced mRNA reads uncovers gene-specific transcription and degradation rates that dynamically change during the cell cycle:

January 17, 2024 at 10:29 AM
Fitting a biophysical model to unspliced and spliced mRNA reads uncovers gene-specific transcription and degradation rates that dynamically change during the cell cycle:
We combined single-cell and single-nucleus sequencing with deep learning and biophysical modelling to analyse single-cell multiome data from mESCs. DeepCycle (doi.org/10.1038/s414...) is effective in sorting both single cells and nuclei according to their cell cycle progression:

January 17, 2024 at 10:28 AM
We combined single-cell and single-nucleus sequencing with deep learning and biophysical modelling to analyse single-cell multiome data from mESCs. DeepCycle (doi.org/10.1038/s414...) is effective in sorting both single cells and nuclei according to their cell cycle progression:
Hey, I am new here! What better way to debut here than to share our recent bioRxiv preprint (the happiest moment during the publication process) on RNA metabolism and chromatin accessibility dynamics during the cell cycle: biorxiv.org/content/10.1... #CellCycle #GeneExpression #SingleCellMultiome

January 17, 2024 at 10:11 AM
Hey, I am new here! What better way to debut here than to share our recent bioRxiv preprint (the happiest moment during the publication process) on RNA metabolism and chromatin accessibility dynamics during the cell cycle: biorxiv.org/content/10.1... #CellCycle #GeneExpression #SingleCellMultiome
New year, new lab preprint! 🎉 I am so happy to share our latest work on RNA metabolism and chromatin accessibility dynamics during the cell cycle in mESCs: https://www.biorxiv.org/content/10.1101/2024.01.11.575159v1
#CellCycle #GeneExpression #SingleCellSequencing #DeepLearning #Biophysics @mauli...
#CellCycle #GeneExpression #SingleCellSequencing #DeepLearning #Biophysics @mauli...

December 6, 2024 at 5:06 PM
New year, new lab preprint! 🎉 I am so happy to share our latest work on RNA metabolism and chromatin accessibility dynamics during the cell cycle in mESCs: https://www.biorxiv.org/content/10.1101/2024.01.11.575159v1
#CellCycle #GeneExpression #SingleCellSequencing #DeepLearning #Biophysics @mauli...
#CellCycle #GeneExpression #SingleCellSequencing #DeepLearning #Biophysics @mauli...
Interested in developing new methods to analyse single-cell sequencing data combining generative deep learning with biophysical principles? Here at the @IGBMC you have a position! #PostdocPosition #ComputationalBiology #SingleCellBiology #DeepLearning #Biophysics (RT 🙏)

December 6, 2024 at 5:06 PM
Interested in developing new methods to analyse single-cell sequencing data combining generative deep learning with biophysical principles? Here at the @IGBMC you have a position! #PostdocPosition #ComputationalBiology #SingleCellBiology #DeepLearning #Biophysics (RT 🙏)
Hey, there are PhD positions on Computational Biophysics available in my lab! Interested? #PhDpositions #ComputationalBiology #ComputationalBiophysics https://jobrxiv.org/job/igbmc-27778-phd-position-on-computational-biophysics/

December 6, 2024 at 5:06 PM
Hey, there are PhD positions on Computational Biophysics available in my lab! Interested? #PhDpositions #ComputationalBiology #ComputationalBiophysics https://jobrxiv.org/job/igbmc-27778-phd-position-on-computational-biophysics/
I lived next to the @CERN for almost five years and never found time to visit it. I have finally resolved this anomaly!


December 6, 2024 at 5:06 PM
I lived next to the @CERN for almost five years and never found time to visit it. I have finally resolved this anomaly!
A great talk from @Naefelix at #EESBioOsc studying circadian rhythms in human from post-mortem samples.

December 6, 2024 at 5:06 PM
A great talk from @Naefelix at #EESBioOsc studying circadian rhythms in human from post-mortem samples.


