
home.kaessmannlab.org
PhD/postdoc positions available - reach out if curious about cerebellum evo-devo and autism spectrum disorders.
We’re based at Uni Tartu, Institute of Genomics (home to Estonian Biobank), and funded by @simonsfoundation.org @embo.org, and the Estonian Research Council.

PhD/postdoc positions available - reach out if curious about cerebellum evo-devo and autism spectrum disorders.
We’re based at Uni Tartu, Institute of Genomics (home to Estonian Biobank), and funded by @simonsfoundation.org @embo.org, and the Estonian Research Council.

PhD/postdoc positions available - reach out if curious about cerebellum evo-devo and autism spectrum disorders.
We’re based at Uni Tartu, Institute of Genomics (home to Estonian Biobank), and funded by @simonsfoundation.org @embo.org, and the Estonian Research Council.
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
@kaessmannlab.bsky.social . Free event, full line up of speakers 👉 www.jeantet.ch/en/
📆Tuesday 14 October, 08h15
📍CMU - Auditoire Alex-F. Müller (A250)
@genevunige.bsky.social

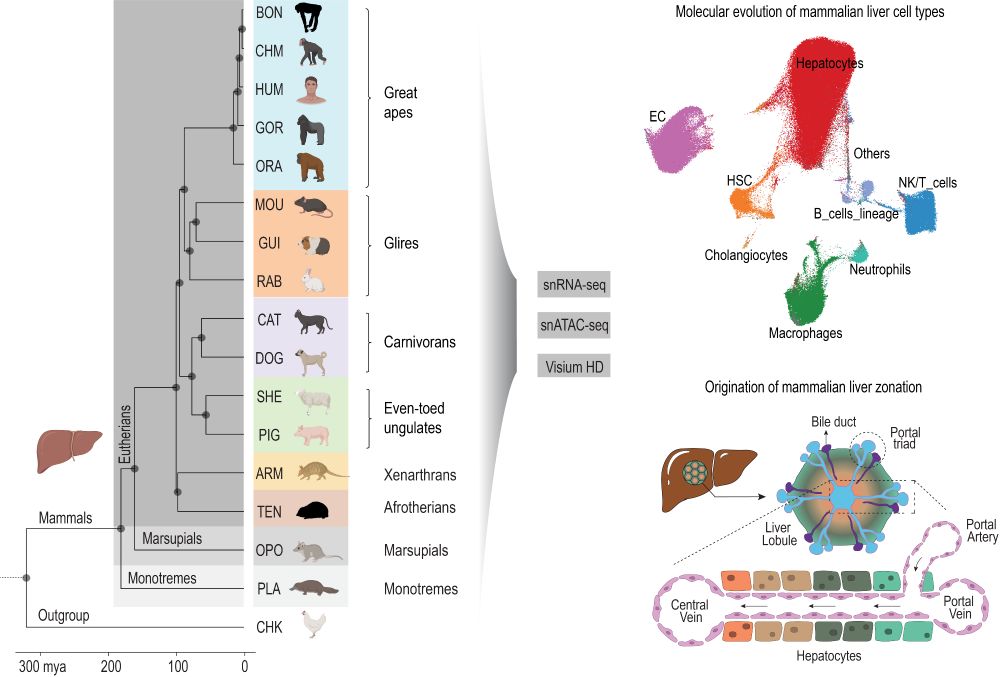
www.nature.com/articles/s41...
www.nature.com/articles/s41...
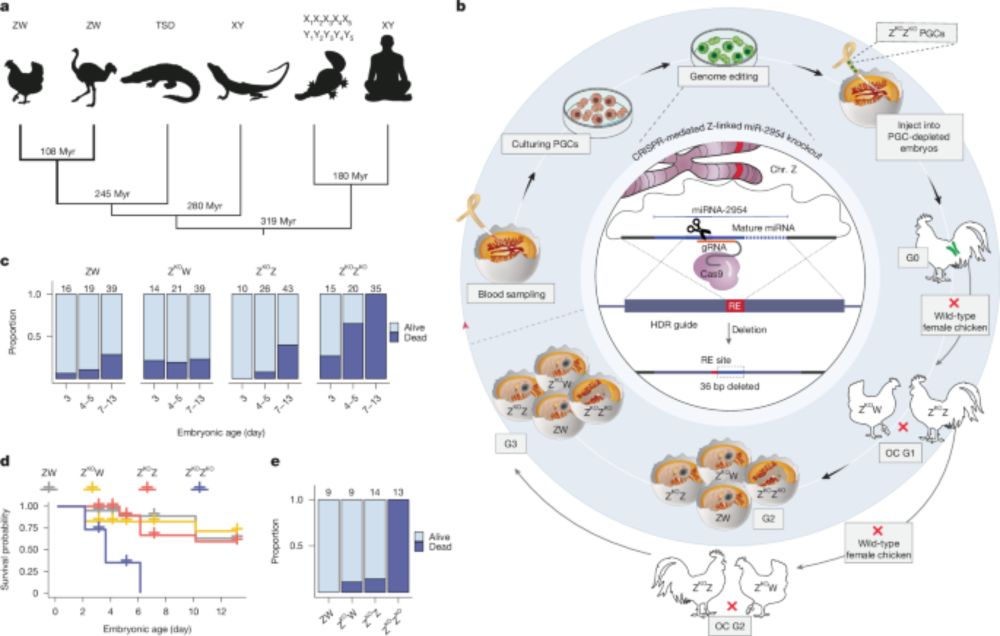
www.uni-heidelberg.de/en/newsroom/...
www.uni-heidelberg.de/en/newsroom/...
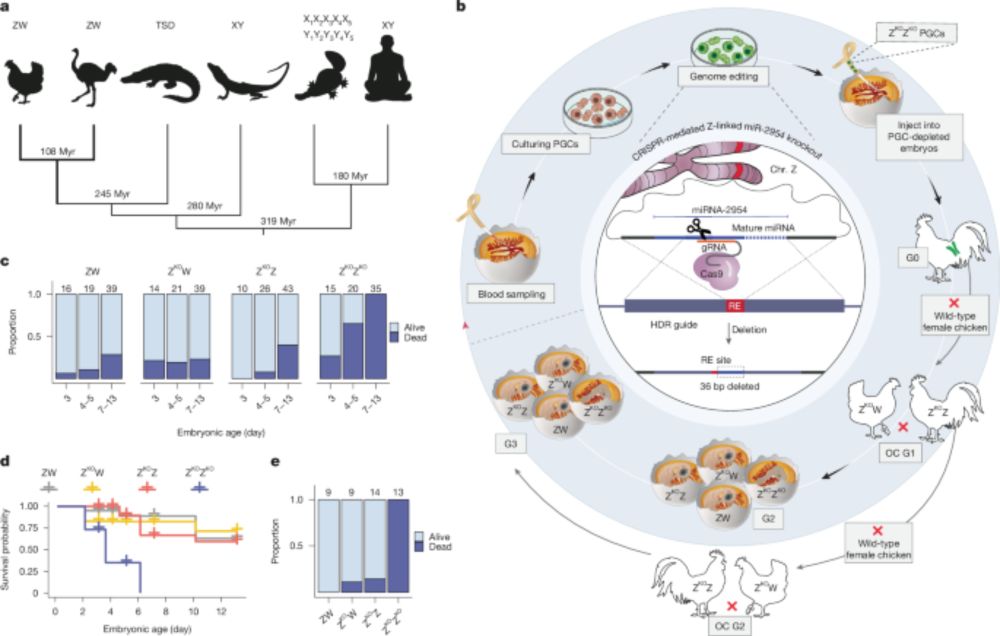
www.biorxiv.org/content/10.1...
Read below for a few highlights...
www.biorxiv.org/content/10.1...
Read below for a few highlights...
www.biorxiv.org/content/10.1...
Read below for a few highlights...
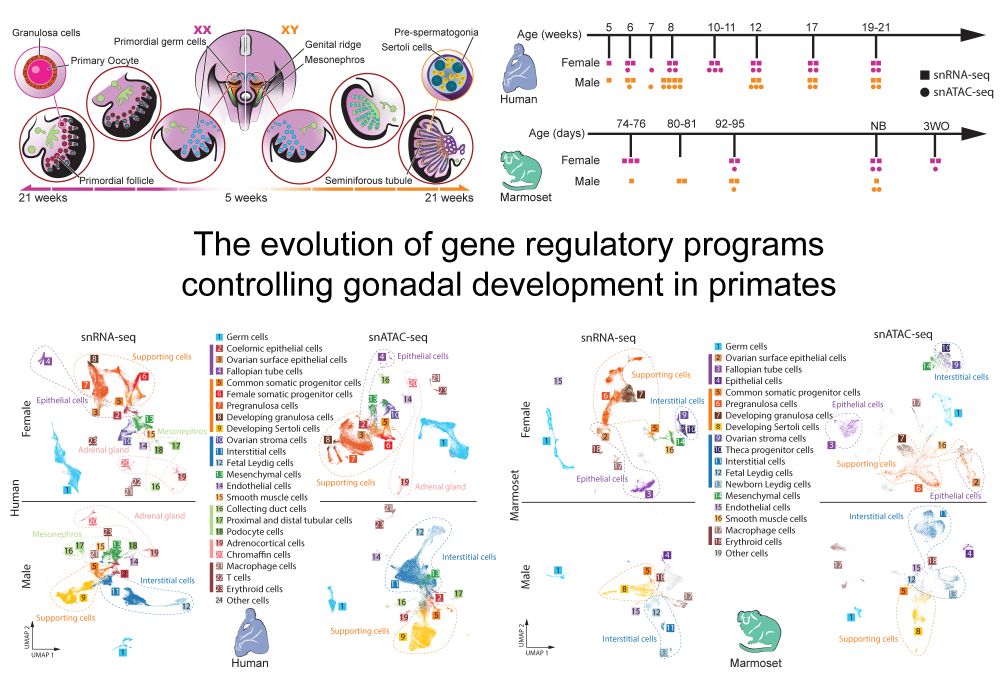

Led by @ioansarr.bsky.social, @marisepp.bsky.social and @tyamadat.bsky.social, in collaboration with @steinaerts.bsky.social
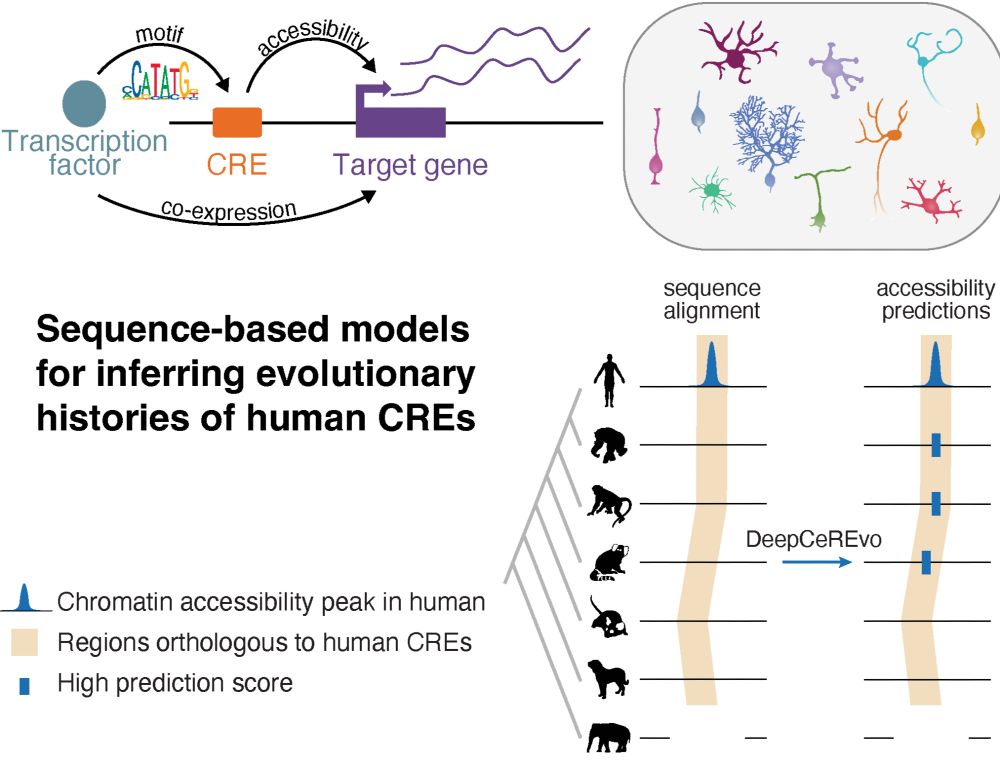
Led by @ioansarr.bsky.social, @marisepp.bsky.social and @tyamadat.bsky.social, in collaboration with @steinaerts.bsky.social

Led by @ioansarr.bsky.social, @marisepp.bsky.social and @tyamadat.bsky.social, in collaboration with @steinaerts.bsky.social
Lots of interesting new TE (& genes) biology
Data fully browsable💻 👉 embryo.helmholtz-munich.de/shiny_embryo/
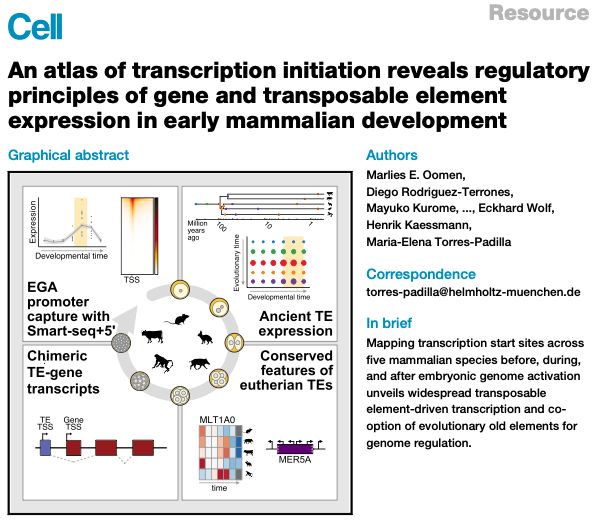
Lots of interesting new TE (& genes) biology
Data fully browsable💻 👉 embryo.helmholtz-munich.de/shiny_embryo/
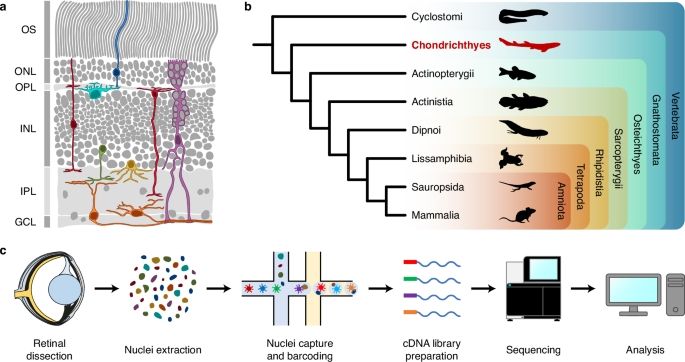
shorturl.at/fvIGZ
I am so incredibly grateful to everyone who made this possible! Especially @kaessmannlab.bsky.social and the García-Moreno lab 💛


shorturl.at/fvIGZ
I am so incredibly grateful to everyone who made this possible! Especially @kaessmannlab.bsky.social and the García-Moreno lab 💛


shorturl.at/fvIGZ
I am so incredibly grateful to everyone who made this possible! Especially @kaessmannlab.bsky.social and the García-Moreno lab 💛

