
Previously PhD @ Sebé-Pedrós lab @crg.eu
Interested in regulatory genomics, evolution, machine learning, and especially the combination of all of the above.
https://anamaria.elek.hr/
www.nature.com/articles/s41...

www.nature.com/articles/s41...
www.nature.com/articles/s41...

www.nature.com/articles/s41...

rdcu.be/eLbaZ

rdcu.be/eLbaZ
🪸 🌊
#evobio #corals #coralbiology
www.nature.com/articles/s41...


🪸 🌊
#evobio #corals #coralbiology
www.nature.com/articles/s41...
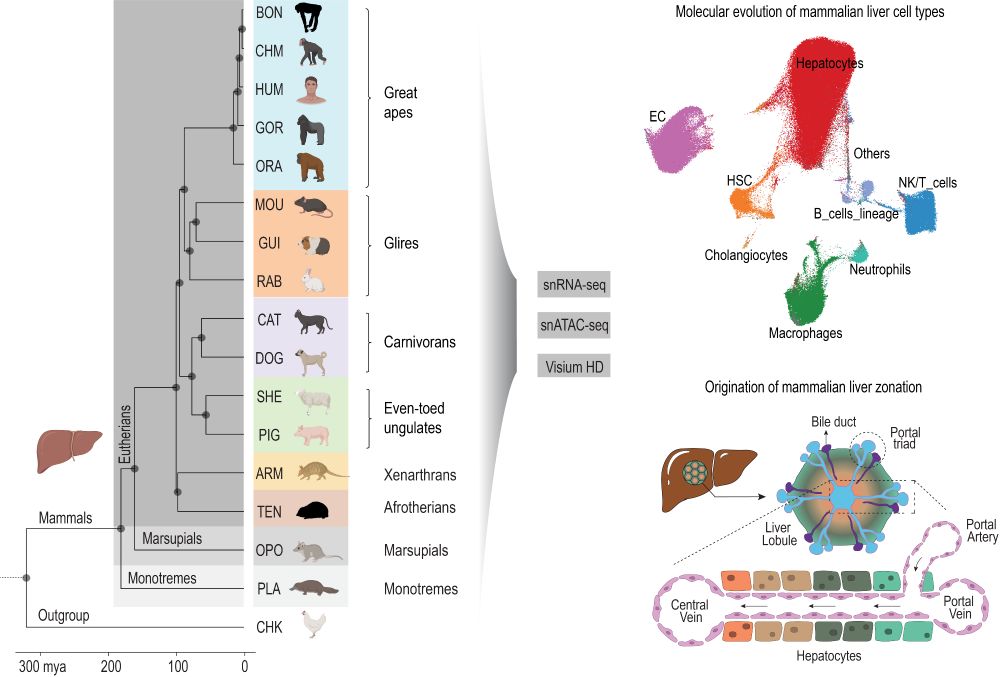
www.nature.com/articles/s41...
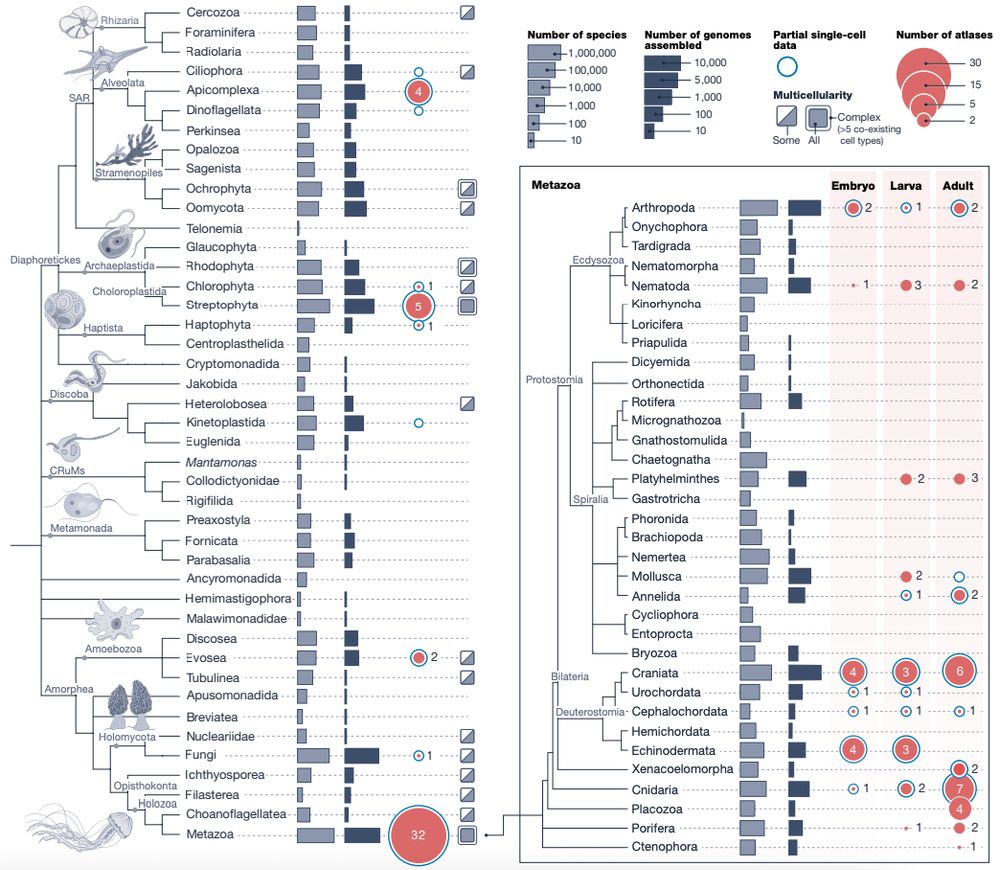
doi.org/10.1146/annu...
Special thanks to @mirimiam.bsky.social, @crg.eu and @upf.edu!

doi.org/10.1146/annu...
Special thanks to @mirimiam.bsky.social, @crg.eu and @upf.edu!
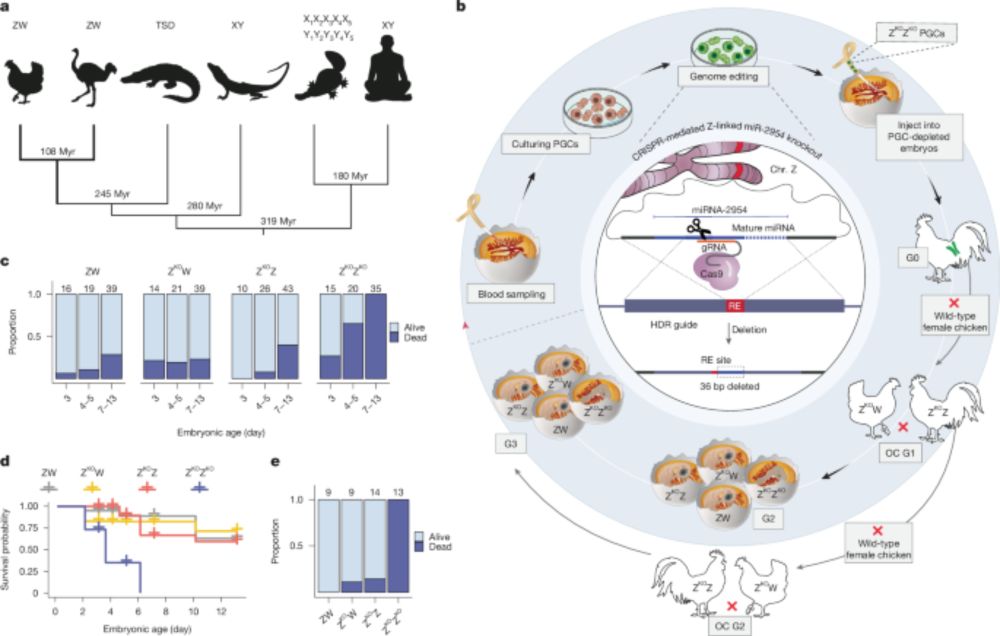
www.biorxiv.org/content/10.1...
Read below for a few highlights...
www.biorxiv.org/content/10.1...
Read below for a few highlights...


We use MPRAs of synthetic enhancers to derive interpretable rules on TFBS arrangement 🚦 and discover that negative synergies drive specificity in hematopoiesis 🩸. Shoutout to @Robert Frömel & @larsplus.bsky.social for leading this work 🦹🦸.
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵

We use MPRAs of synthetic enhancers to derive interpretable rules on TFBS arrangement 🚦 and discover that negative synergies drive specificity in hematopoiesis 🩸. Shoutout to @Robert Frömel & @larsplus.bsky.social for leading this work 🦹🦸.
🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵

🧬🩸 screen of fully synthetic enhancers in blood progenitors
🤖 AI that creates new cell state specific enhancers
🔍 negative synergies between TFs lead to specificity!
www.cell.com/cell/fulltex...
🧵

In our new review, @fedemantica.bsky.social and I argue we are missing the most prevalent one: specialization. And the same applies to alternative splicing! 1/7
tinyurl.com/45k7kbmp
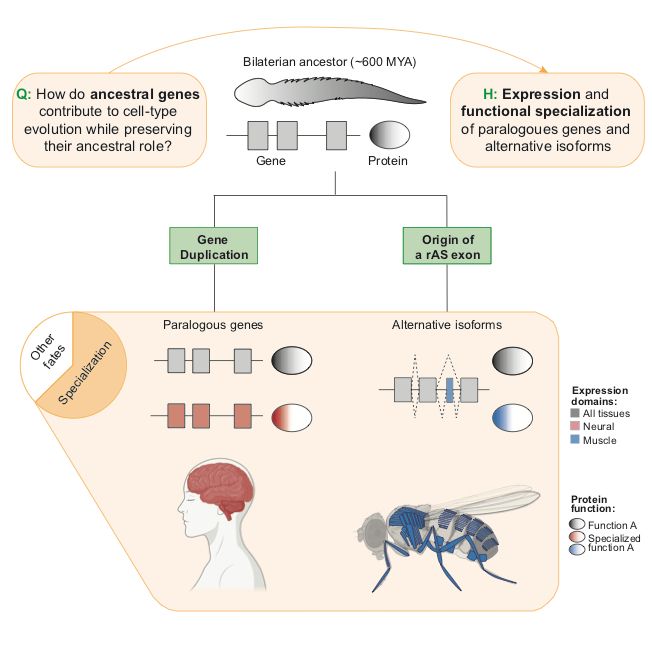
In our new review, @fedemantica.bsky.social and I argue we are missing the most prevalent one: specialization. And the same applies to alternative splicing! 1/7
tinyurl.com/45k7kbmp
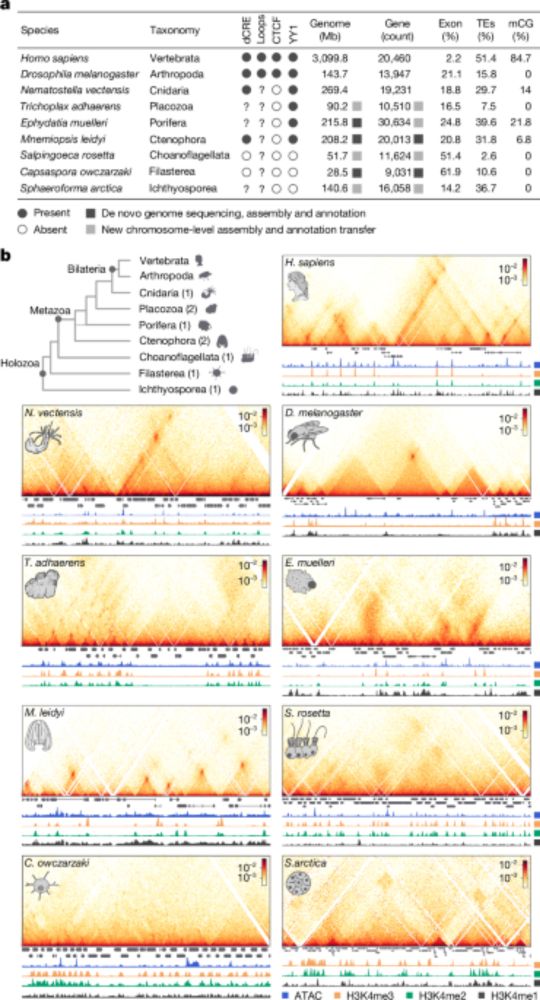
Here @crisnava.bsky.social, @seanamontgomery.bsky.social & collaborators develop a novel ChIPseq protocol, and demonstrate its huge potential to study the evolution of chromatin function and regulation across the eukaryotic tree of life.

Here @crisnava.bsky.social, @seanamontgomery.bsky.social & collaborators develop a novel ChIPseq protocol, and demonstrate its huge potential to study the evolution of chromatin function and regulation across the eukaryotic tree of life.

