

go.nature.com/4pAbARf

go.nature.com/4pAbARf
www.quantamagazine.org/loops-of-dna...

www.quantamagazine.org/loops-of-dna...

These five websites offer free scientific illustrations for biologists. Great for presentations, research papers and other research communication needs.
Save and share the post!

These five websites offer free scientific illustrations for biologists. Great for presentations, research papers and other research communication needs.
Save and share the post!
with #LauraPiovani @dariagavr.bsky.social @alexdemendoza.bsky.social @chemamd.bsky.social and others /1

with #LauraPiovani @dariagavr.bsky.social @alexdemendoza.bsky.social @chemamd.bsky.social and others /1
authors.elsevier.com/sd/article/S...
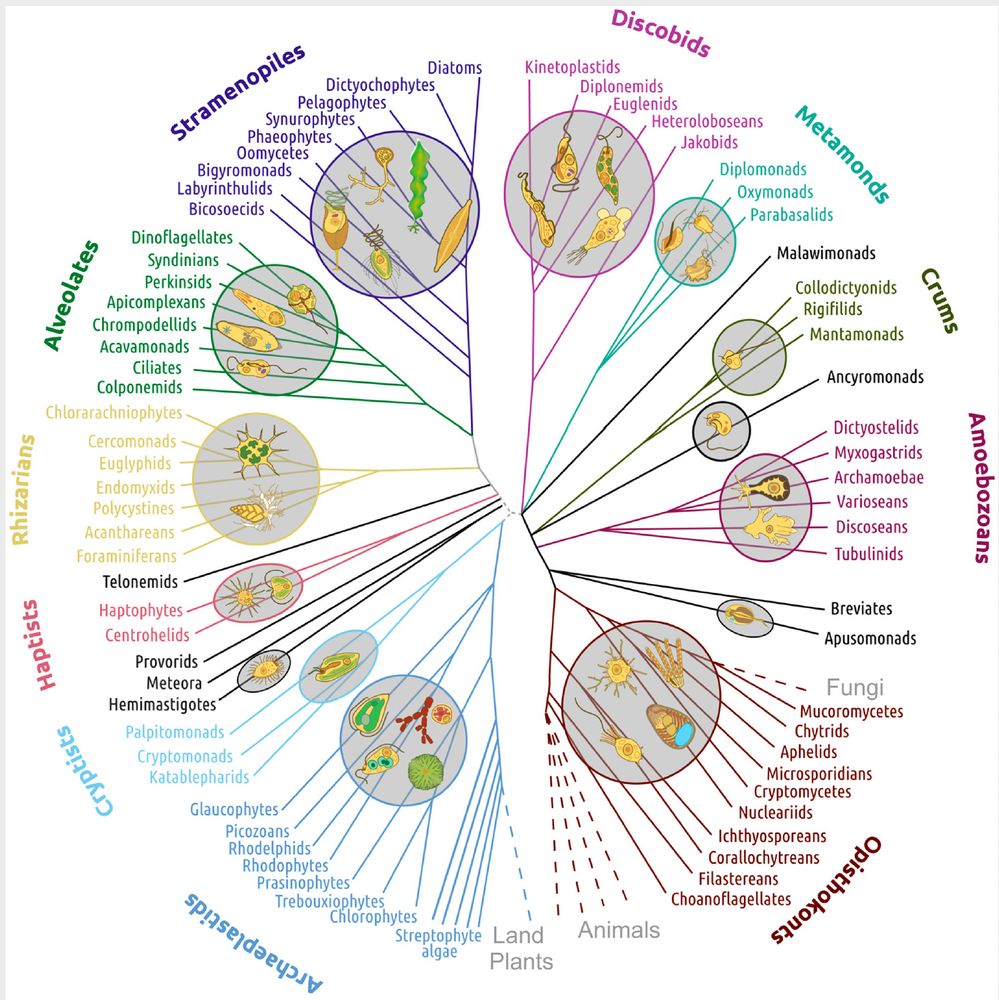
Check out our paper - fresh off the press!!!
We find widespread functional conservation of enhancers in absence of sequence homology
Including: a bioinformatic tool to map sequence-diverged enhancers!
rdcu.be/enVDN
github.com/tobiaszehnde...

Check out our paper - fresh off the press!!!
We find widespread functional conservation of enhancers in absence of sequence homology
Including: a bioinformatic tool to map sequence-diverged enhancers!
rdcu.be/enVDN
github.com/tobiaszehnde...
www.science.org/doi/10.1126/...
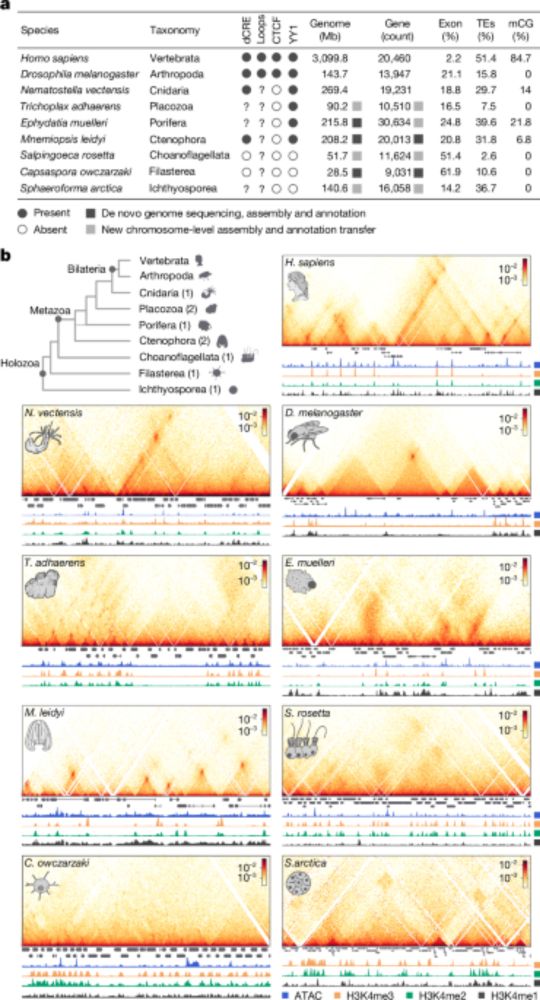
🧬Our Repli-Histo labeling marks nucleosomes in euchromatin and heterochromatin in live human cells.
🔍 @katsuminami.bsky.social et al. have developed a chromatin behavior atlas within the nucleus. 1/2
www.science.org/doi/10.1126/...
🧬Our Repli-Histo labeling marks nucleosomes in euchromatin and heterochromatin in live human cells.
🔍 @katsuminami.bsky.social et al. have developed a chromatin behavior atlas within the nucleus. 1/2
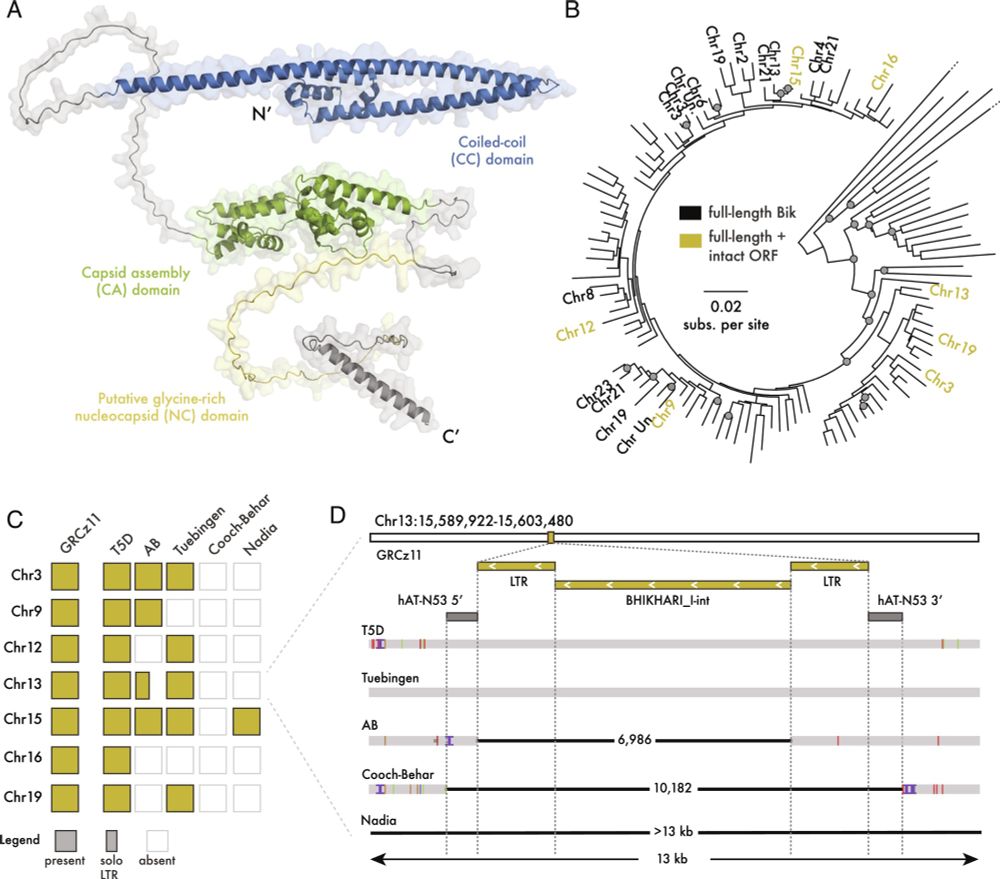
Gag proteins of endogenous retroviruses are required for zebrafish development
www.pnas.org/doi/10.1073/...
Led heroically by Sylvia Chang & @jonowells.bsky.social
A study which has changed the way I think of #transposons! No less! 🧵 1/n
Gag proteins of endogenous retroviruses are required for zebrafish development
www.pnas.org/doi/10.1073/...
Led heroically by Sylvia Chang & @jonowells.bsky.social
A study which has changed the way I think of #transposons! No less! 🧵 1/n
www.science.org/doi/10.1126/...
www.sciencedirect.com/science/arti...
www.sciencedirect.com/science/arti...
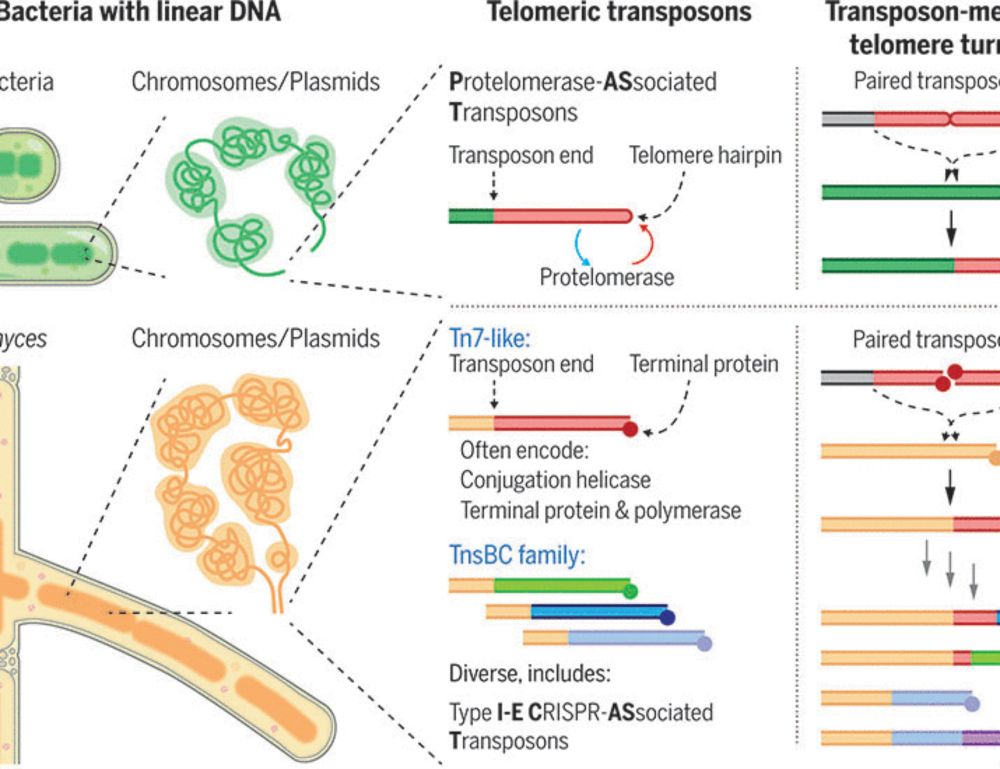
www.science.org/doi/10.1126/...
www.sciencedirect.com/science/arti...
www.sciencedirect.com/science/arti...


A 🧵 below 👇🏼





sciencedirect.com/science/arti...
A big thank you to Tom Fillot for his efforts on this and to
@hansen_lab
@marcelonollmann
for their help as editors.
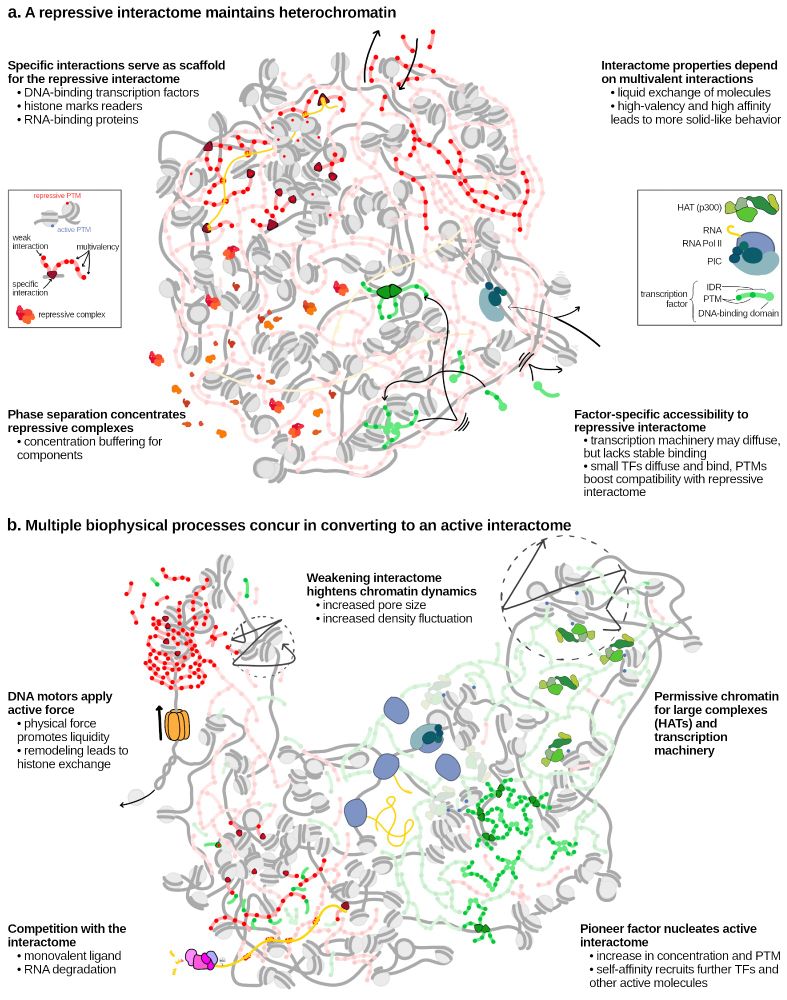
sciencedirect.com/science/arti...
A big thank you to Tom Fillot for his efforts on this and to
@hansen_lab
@marcelonollmann
for their help as editors.
We are seeking candidates with a background in computational and/or molecular biology, and with a broad interest in evolutionary biology, gene regulation, and comparative genomics.
📅Apply before January 12 here: www.crg.eu/en/content/t...
We are seeking candidates with a background in computational and/or molecular biology, and with a broad interest in evolutionary biology, gene regulation, and comparative genomics.
📅Apply before January 12 here: www.crg.eu/en/content/t...
www.mghlab.org/software

www.mghlab.org/software
www.cell.com/action/showP...
www.cell.com/action/showP...

