
📄 Paper: www.nature.com/articles/s41...
💻 Weights: github.com/google-deepm...
Getting here wasn’t a straight path. We discussed the story behind the model, paper & API in the following roundtable: youtu.be/V8lhUqKqzUc

📄 Paper: www.nature.com/articles/s41...
💻 Weights: github.com/google-deepm...
Getting here wasn’t a straight path. We discussed the story behind the model, paper & API in the following roundtable: youtu.be/V8lhUqKqzUc
"Better late than never?" the source says.
That's because crucial NIH grant-review panels are slated to be empty at those institutes by Jan 2027.
A wonky bureaucratic problem with big implications.
A short 🧵

"Better late than never?" the source says.
Children shouldn't be locked up by our government.

Children shouldn't be locked up by our government.
talkingpointsmemo.com/news/democra...

talkingpointsmemo.com/news/democra...
go.nature.com/4qu2ExZ

go.nature.com/4qu2ExZ
We use them to map 665,856 pairwise genetic perturbations and outline a path to comprehensive interaction mapping in human cells.
We also introduce an approach for cloning lentiviral libraries with billions of elements.

We use them to map 665,856 pairwise genetic perturbations and outline a path to comprehensive interaction mapping in human cells.
We also introduce an approach for cloning lentiviral libraries with billions of elements.
We mapped the regulatory landscape of human liver at single-nucleus resolution using snMultiome, connecting genetic variants to the cell-type mechanisms underlying cardiometabolic disease.
www.sciencedirect.com/science/arti...

We mapped the regulatory landscape of human liver at single-nucleus resolution using snMultiome, connecting genetic variants to the cell-type mechanisms underlying cardiometabolic disease.
www.sciencedirect.com/science/arti...
www.appropriations.senate.gov/news/majorit...
1/5

www.appropriations.senate.gov/news/majorit...
1/5
Together with @jengreitz.bsky.social, Howard Chang, and team, we found that answering this question is much more challenging than previously thought!
academic.oup.com/nar/article/...
See thread below:

Together with @jengreitz.bsky.social, Howard Chang, and team, we found that answering this question is much more challenging than previously thought!
academic.oup.com/nar/article/...
See thread below:
TLDR; TFIID behaves differently depending on promoter type. More below:

TLDR; TFIID behaves differently depending on promoter type. More below:
Think again!
Thread coming soon

Think again!
Thread coming soon
In a new pre-print, we find that TWAS gets the sign wrong around 20-30% of the time!
doi.org/10.64898/202...
1/n

In a new pre-print, we find that TWAS gets the sign wrong around 20-30% of the time!
doi.org/10.64898/202...
1/n

@moffittlab.bsky.social, @saramostafavi.bsky.social, me and all speakers for the 2026 CSHL meeting Systems Biology: Global Regulation of Gene Expression, March 11-14. Abstract deadline January 9! More infos and registration at meetings.cshl.edu/meetings.asp...

@moffittlab.bsky.social, @saramostafavi.bsky.social, me and all speakers for the 2026 CSHL meeting Systems Biology: Global Regulation of Gene Expression, March 11-14. Abstract deadline January 9! More infos and registration at meetings.cshl.edu/meetings.asp...

Despite driving ~2/3 of mammalian genes, CpG island (CGI) promoters have remained a puzzle. We identified >50 activators that are exclusively compatible with this promoter class. 🧬
Despite driving ~2/3 of mammalian genes, CpG island (CGI) promoters have remained a puzzle. We identified >50 activators that are exclusively compatible with this promoter class. 🧬
nature.com/articles/s41...
1/10

nature.com/articles/s41...
1/10

Our latest work on causal models for this is out yesterday:
www.nature.com/articles/s41...
A short🧵:

Our latest work on causal models for this is out yesterday:
www.nature.com/articles/s41...
A short🧵:


Here’s my most recent one covering the latest findings on ecDNA retention mechanisms from the Chang & Mischel labs.
Here’s my most recent one covering the latest findings on ecDNA retention mechanisms from the Chang & Mischel labs.
We profiled ~10M PBMCs (snRNA-seq + snATAC-seq) from 1,108 Finnish donors to map how genetic variants drive complex disease through chromatin and gene regulation 🧵👇
🔗 Link: www.medrxiv.org/content/10.1...
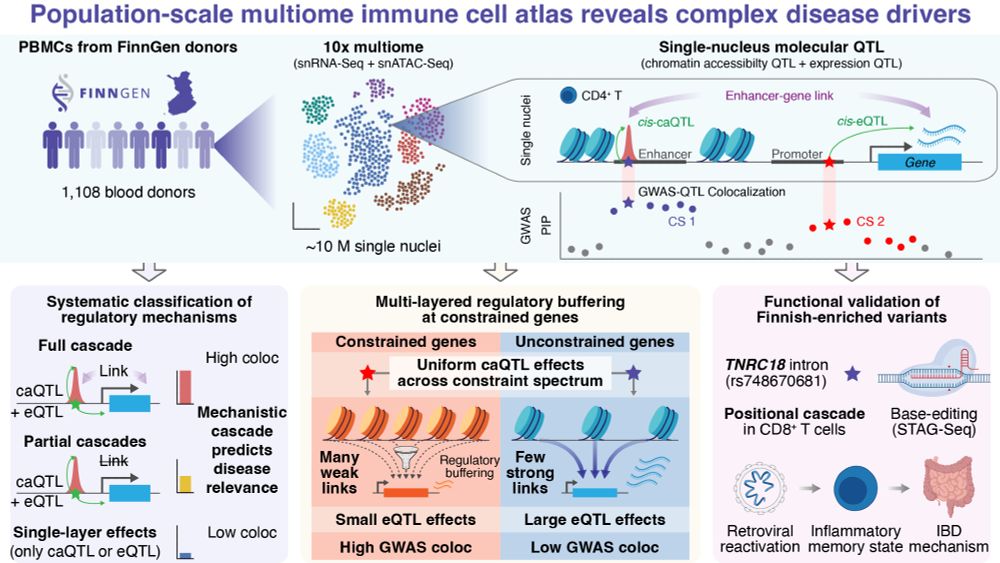
We profiled ~10M PBMCs (snRNA-seq + snATAC-seq) from 1,108 Finnish donors to map how genetic variants drive complex disease through chromatin and gene regulation 🧵👇
🔗 Link: www.medrxiv.org/content/10.1...
1/n

