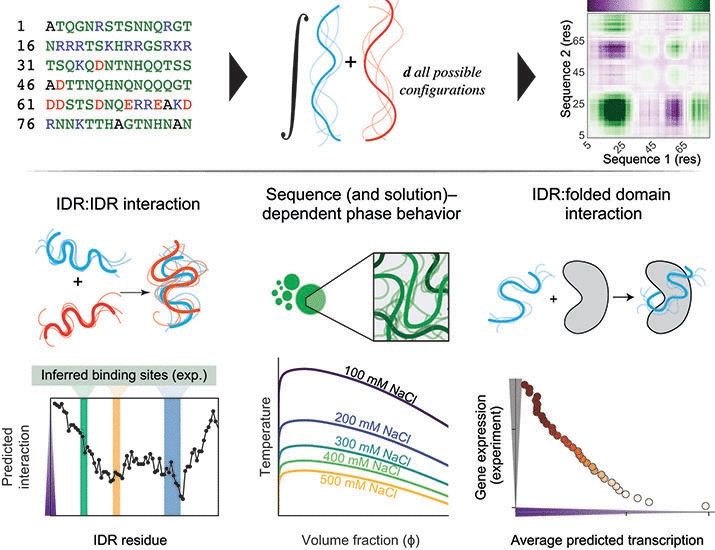
www.biorxiv.org/content/10.1...


Remaining talks to be filled from submissions - register & submit by 04 Sep 2025 to join us in Mexico.
Speakers, details & registration➡️ bit.ly/46JQbze


Remaining talks to be filled from submissions - register & submit by 04 Sep 2025 to join us in Mexico.
Speakers, details & registration➡️ bit.ly/46JQbze
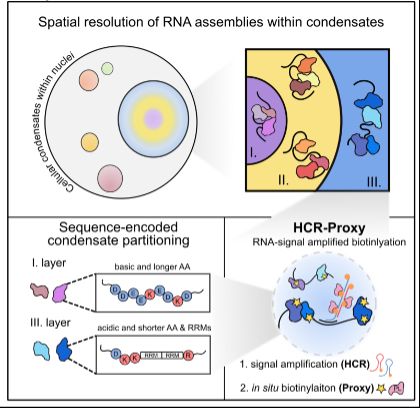
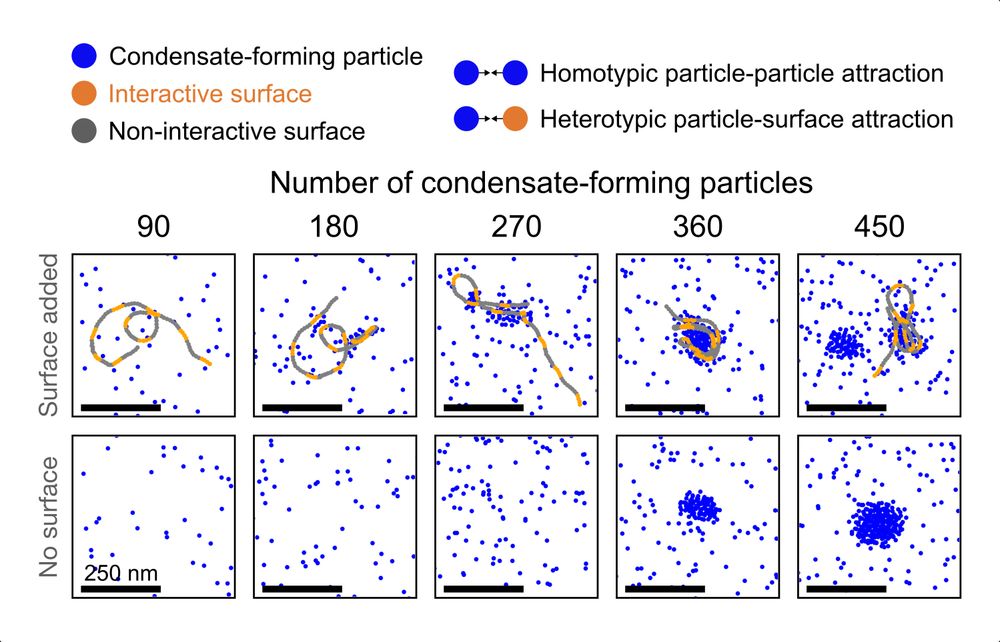
HCR-Proxy, a modular proximity labelling approach to profile local proteome composition around RNA at nanoscale, subcompartmental resolution. Thereby we resolved nested nucleolar layers and uncovered the grammar of spatial protein partitioning.
www.biorxiv.org/content/10.1...

www.pse.kit.edu/english/karr...
www.pse.kit.edu/english/karr...
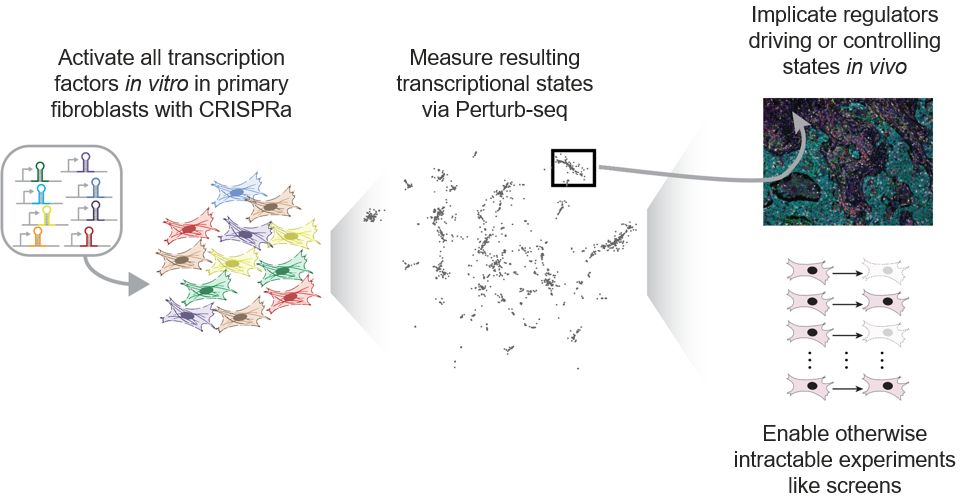
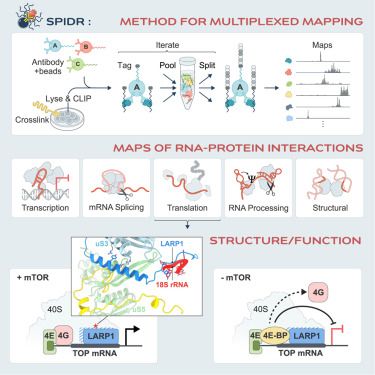
www.cell.com/cell/fulltex...
Join us, if RNP assembly on centromeres, RNA modifications, & bioinformatics are your thing.
More info also on modiclab.org.
The project is in collaboration with my new collegue Miha Modic. If you are interesed in RNA modification, centromeres and bioinformatics this is for you!
Miha has more positions to fill, infos on his linkedin: linkedin.com/in/miha-modic-9aa8a752.

Join us, if RNP assembly on centromeres, RNA modifications, & bioinformatics are your thing.
More info also on modiclab.org.
HCR-Proxy, a modular proximity labelling approach to profile local proteome composition around RNA at nanoscale, subcompartmental resolution. Thereby we resolved nested nucleolar layers and uncovered the grammar of spatial protein partitioning.
www.biorxiv.org/content/10.1...

HCR-Proxy, a modular proximity labelling approach to profile local proteome composition around RNA at nanoscale, subcompartmental resolution. Thereby we resolved nested nucleolar layers and uncovered the grammar of spatial protein partitioning.
www.biorxiv.org/content/10.1...
1/6
1/6
Welcome new T-Track Prof. Miha Modic @paraspeckle.bsky.social to @kit.edu Karlsruhe
I am so very excited to have you with us now!

Here is the link that works! authors.elsevier.com/a/1kpxo_Oylr...
Here is the link that works! authors.elsevier.com/a/1kpxo_Oylr...
podcasts.apple.com/us/podcast/n... @nightsciencepod.bsky.social

podcasts.apple.com/us/podcast/n... @nightsciencepod.bsky.social

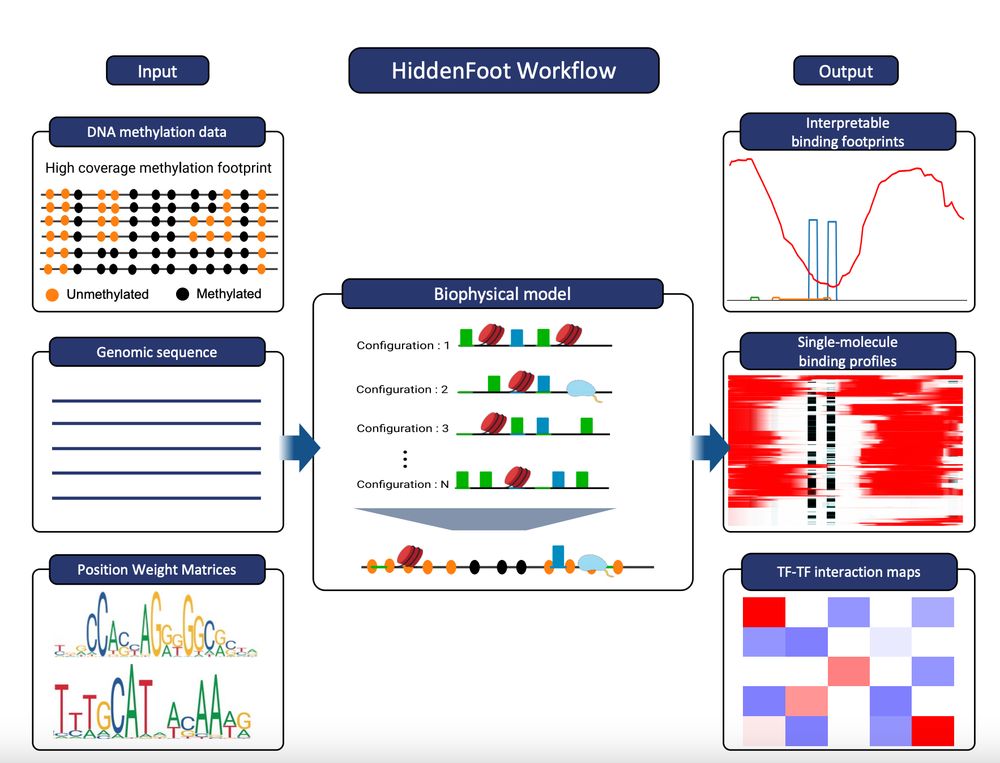
genomebiology.biomedcentral.com/articles/10....
genomebiology.biomedcentral.com/articles/10....

shorturl.at/pmLIj #RNASky

shorturl.at/pmLIj #RNASky