
Previously at @saezlab.bsky.social
Core developer of https://decoupler.readthedocs.io/ at @scverse.bsky.social
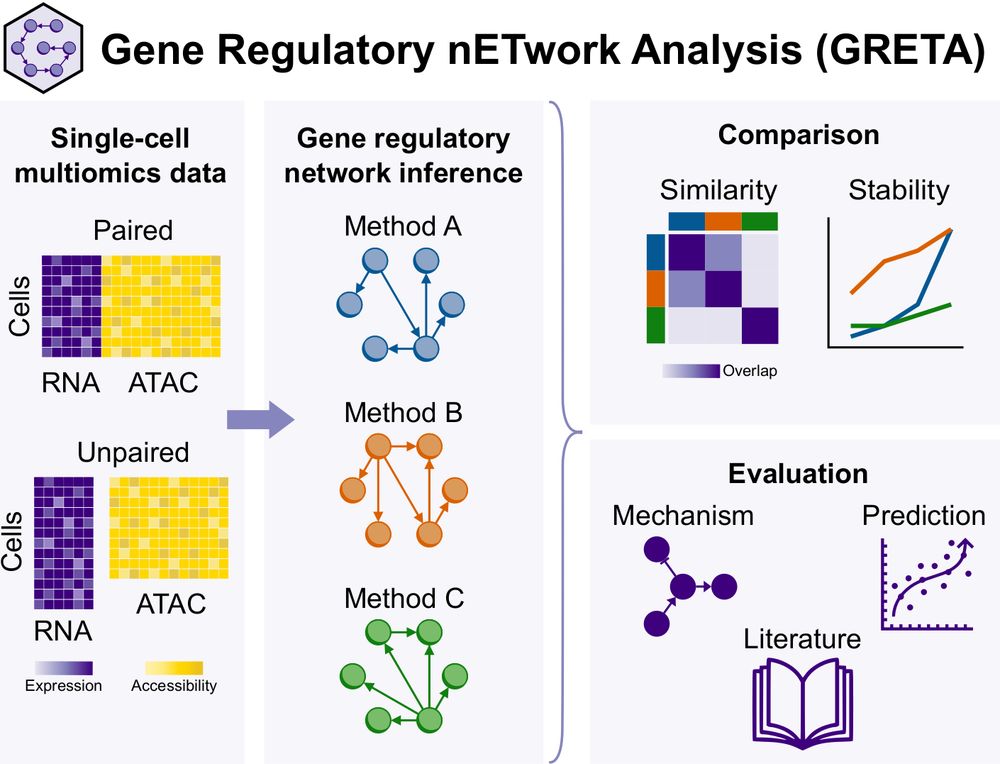
Paper: doi.org/10.1101/2024.12.20.629764
Code: github.com/saezlab/greta



@twangmdphd.bsky.social ! CRISPR base editors are driving several clinical trials, but it's hard to quantify off-target edits and their effects. Tong developed a sequencing assay to identify off-targets *in primary cells* 🧬🖥️🧪 #genomics #chromatin #CRISPR
www.biorxiv.org/content/10.1...

@twangmdphd.bsky.social ! CRISPR base editors are driving several clinical trials, but it's hard to quantify off-target edits and their effects. Tong developed a sequencing assay to identify off-targets *in primary cells* 🧬🖥️🧪 #genomics #chromatin #CRISPR
Remote attendance is available for 30 USD, and you can secure your spot by registering at www.eventbrite.com/e/scverse-co....
We hope to see you online at the conference!

I uploaded the classic Watson & Crick paper about DNA structure, and the Adviser had this to say about one of the greatest paper endings of the century:

I uploaded the classic Watson & Crick paper about DNA structure, and the Adviser had this to say about one of the greatest paper endings of the century:


Excited to share SEAM: Systematic Explanation of Attribtuion-based Mechanisms. SEAM is an explainable AI method that dissects cis-regulatory mechanisms learned by seq2fun genomic deep learning models.
Led by @EESetiz
1/N 🧵👇

Excited to share SEAM: Systematic Explanation of Attribtuion-based Mechanisms. SEAM is an explainable AI method that dissects cis-regulatory mechanisms learned by seq2fun genomic deep learning models.
Led by @EESetiz
1/N 🧵👇
We're proud to recognize Stellaromics as a Gold Sponsor of scverse conference 2025! 🥇
@stellaromics.bsky.social
#scverse2025 #SpatialBiology #3DSpatial #ComputationalBiology #SingleCell #Stellaromics #SpatialOmics #Bioinformatics


We're proud to recognize Stellaromics as a Gold Sponsor of scverse conference 2025! 🥇
@stellaromics.bsky.social
#scverse2025 #SpatialBiology #3DSpatial #ComputationalBiology #SingleCell #Stellaromics #SpatialOmics #Bioinformatics
The goal of my lab is to understand the regulatory role of every nucleotide in our genomes and how this changes across every cell in our bodies.
The goal of my lab is to understand the regulatory role of every nucleotide in our genomes and how this changes across every cell in our bodies.
www.quantamagazine.org/loops-of-dna...

www.quantamagazine.org/loops-of-dna...
Processing and Modeling Large-Scale Single-Cell Perturbation Data with Machine Learning
🧵
@arcinstitute.org
#scverse2025 #PerturbSeq #MachineLearning #SingleCell #CRISPR #GenomicScreening #VirtualCells

Processing and Modeling Large-Scale Single-Cell Perturbation Data with Machine Learning
🧵
@arcinstitute.org
#scverse2025 #PerturbSeq #MachineLearning #SingleCell #CRISPR #GenomicScreening #VirtualCells
In 2020, Goodall spoke with Science Editor-in-Chief H. Holden Thorp for a #ScienceEditorial reflecting on the 60th anniversary of her arrival in Gombe to study wild chimpanzees. Read more: https://scim.ag/472Chb8

In 2020, Goodall spoke with Science Editor-in-Chief H. Holden Thorp for a #ScienceEditorial reflecting on the 60th anniversary of her arrival in Gombe to study wild chimpanzees. Read more: https://scim.ag/472Chb8
🔗 Manuscript: www.biorxiv.org/content/10.1...
💻 Code: partipy.readthedocs.io

🔗 Manuscript: www.biorxiv.org/content/10.1...
💻 Code: partipy.readthedocs.io
Submissions and travel grant applications close in 3 days. Find out more and apply at scverse.org/conference2025
@scverse.bsky.social

Submissions and travel grant applications close in 3 days. Find out more and apply at scverse.org/conference2025
@scverse.bsky.social
Explain your answer.
Explain your answer.
CREsted allows you to train sequence-to-function models tailored to cell type-specific enhancers (including synthetic enhancer design) www.biorxiv.org/content/10.1... github.com/aertslab/cre...
(Zoom registration link and more information in thread!)
🧵
CREsted allows you to train sequence-to-function models tailored to cell type-specific enhancers (including synthetic enhancer design) www.biorxiv.org/content/10.1... github.com/aertslab/cre...

embl.wd103.myworkdayjobs.com/en-US/EMBL/j...
(1/3)

embl.wd103.myworkdayjobs.com/en-US/EMBL/j...
(1/3)


www.nature.com/articles/s41...

www.nature.com/articles/s41...
A story of patience, absurdity, and persistence 🌀 <1min
From Alfonso Valencia’s lab and a very stubborn PhD student (me).

🧬 64% of disease co-occurrences can be explained by transcriptomic similarities.
Comorbidities aren’t random—they have a molecular basis.
Here’s how we found it 👇 (1/n)
🔗 doi.org/10.1073/pnas.2421060122
@alfonsovalencia.bsky.social

🧬 64% of disease co-occurrences can be explained by transcriptomic similarities.
Comorbidities aren’t random—they have a molecular basis.
Here’s how we found it 👇 (1/n)
🔗 doi.org/10.1073/pnas.2421060122
@alfonsovalencia.bsky.social
A story of patience, absurdity, and persistence 🌀 <1min
From Alfonso Valencia’s lab and a very stubborn PhD student (me).

A story of patience, absurdity, and persistence 🌀 <1min
From Alfonso Valencia’s lab and a very stubborn PhD student (me).

