
We also presented our latest findings from the lab 👇

We also presented our latest findings from the lab 👇
- Clear metadata for annotations
- Incentivise the production of annotated datasets
- Reduce the number of formats used for annotations
- Make data accessible to all
Huge thanks to all collaborators—can’t wait to see MIFA in action!
- Clear metadata for annotations
- Incentivise the production of annotated datasets
- Reduce the number of formats used for annotations
- Make data accessible to all
Huge thanks to all collaborators—can’t wait to see MIFA in action!
Submissions and travel grant applications close in 3 days. Find out more and apply at scverse.org/conference2025
@scverse.bsky.social

Submissions and travel grant applications close in 3 days. Find out more and apply at scverse.org/conference2025
@scverse.bsky.social
Did you know we are offering grants to help anyone in financial need attend our annual conference? 🌍
🧵
#scverse #scverse2025 #SingleCell #Conference #StanfordUniversity #TravelGrant

Did you know we are offering grants to help anyone in financial need attend our annual conference? 🌍
🧵
#scverse #scverse2025 #SingleCell #Conference #StanfordUniversity #TravelGrant
- OMERO-vitessce
- IO-FAST initiative
- @nfdi4bioimage.bsky.social Data Stewardship
Talks, Workshops & Poster Session 2 up ahead today!🔬
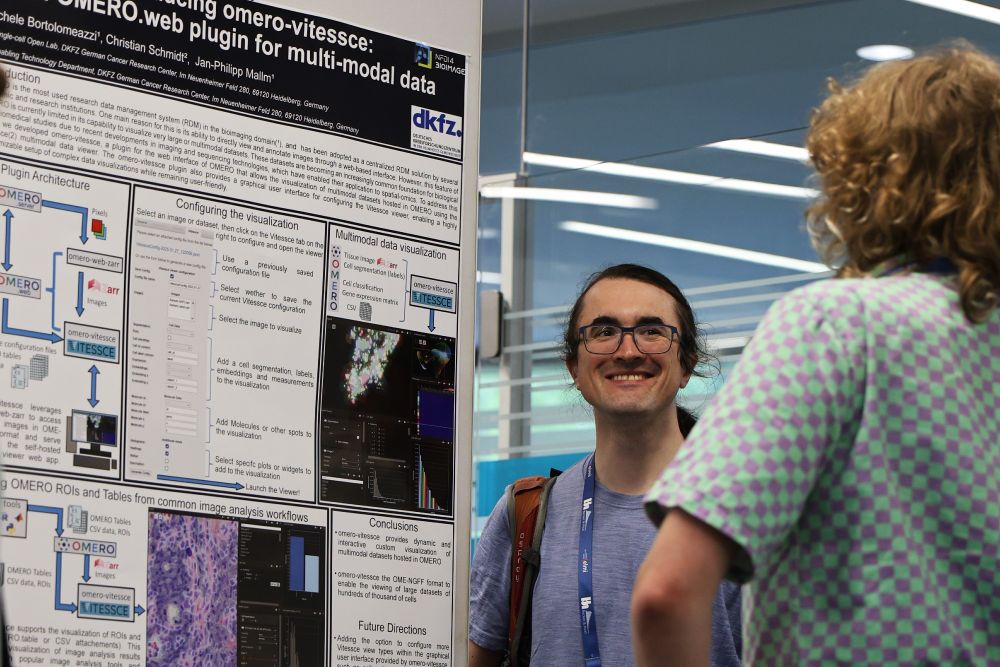



- OMERO-vitessce
- IO-FAST initiative
- @nfdi4bioimage.bsky.social Data Stewardship
Talks, Workshops & Poster Session 2 up ahead today!🔬
Call for abstracts and registrations coming soon!

Call for abstracts and registrations coming soon!

Segger provides accurate and scalable segmentation for IIS and FISH-based spatial omics datasets, using a novel heterogenous GNN approach.
Authors: elihei.bsky.social and Andrew Moorman.
Segger provides accurate and scalable segmentation for IIS and FISH-based spatial omics datasets, using a novel heterogenous GNN approach.
Authors: elihei.bsky.social and Andrew Moorman.
Our report on the 2024 OME-NGFF Workflows Hackathon is a deep dive into the status of OME-Zarr and the great work that happened last November at the BioVisionCenter in Zurich.
🔗 Read the preprint: osf.io/preprints/bi...
🧵 (1/15)
Our report on the 2024 OME-NGFF Workflows Hackathon is a deep dive into the status of OME-Zarr and the great work that happened last November at the BioVisionCenter in Zurich.
🔗 Read the preprint: osf.io/preprints/bi...
🧵 (1/15)
Looking forward to the new in-person event!



Looking forward to the new in-person event!







