
Check it out here: www.biorxiv.org/content/10.6...
Congratulations and thanks to everyone involved for the great effort!

Check it out here: www.biorxiv.org/content/10.6...
Congratulations and thanks to everyone involved for the great effort!
arxiv.org/abs/2512.12272

arxiv.org/abs/2512.12272
We introduce AbTags – antibody–peptide fusion proteins that massively amplify signal, enable absolute quantification of binding events, and are read out together with the single-cell proteome by LC–MS.
AbTag platform is patent-pending.
Preprint: www.biorxiv.org/content/10.6...

We introduce AbTags – antibody–peptide fusion proteins that massively amplify signal, enable absolute quantification of binding events, and are read out together with the single-cell proteome by LC–MS.
AbTag platform is patent-pending.
Preprint: www.biorxiv.org/content/10.6...
We can now give clear guidance on what tests perform well. Thank you Tjorven Hinzke and
@benoitkunath.bsky.social for the leadership

We can now give clear guidance on what tests perform well. Thank you Tjorven Hinzke and
@benoitkunath.bsky.social for the leadership
www.biorxiv.org/content/10.6...

www.biorxiv.org/content/10.6...
arxiv.org/abs/2512.20954

arxiv.org/abs/2512.20954





Many thanks to @beilstein-institut.bsky.social @glycosmos.bsky.social @sib.swiss

Many thanks to @beilstein-institut.bsky.social @glycosmos.bsky.social @sib.swiss

Huge thanks to everyone, and special thanks to @stephanhacker2.bsky.social and @pzanon.bsky.social
Our study on "Profiling the proteome-wide selectivity of diverse electrophiles" is published in Nature Chemistry.(1/7)
www.nature.com/articles/s41...

Huge thanks to everyone, and special thanks to @stephanhacker2.bsky.social and @pzanon.bsky.social



openrxiv.org/enabling-rev...

Thanks to @willfondrie.com for alerting me to this amazing repo.
genzplyr is dplyr, but bussin fr fr no cap.

Thanks to @willfondrie.com for alerting me to this amazing repo.

A collaboration to give the Flaviviridae (home to Zika, Dengue & HCV) a much-needed taxonomic re-think.
Our at-scale AI structure prediction gave a complementary perspective on viral evolution.
www.nature.com/articles/s41...

A collaboration to give the Flaviviridae (home to Zika, Dengue & HCV) a much-needed taxonomic re-think.
Our at-scale AI structure prediction gave a complementary perspective on viral evolution.
www.nature.com/articles/s41...
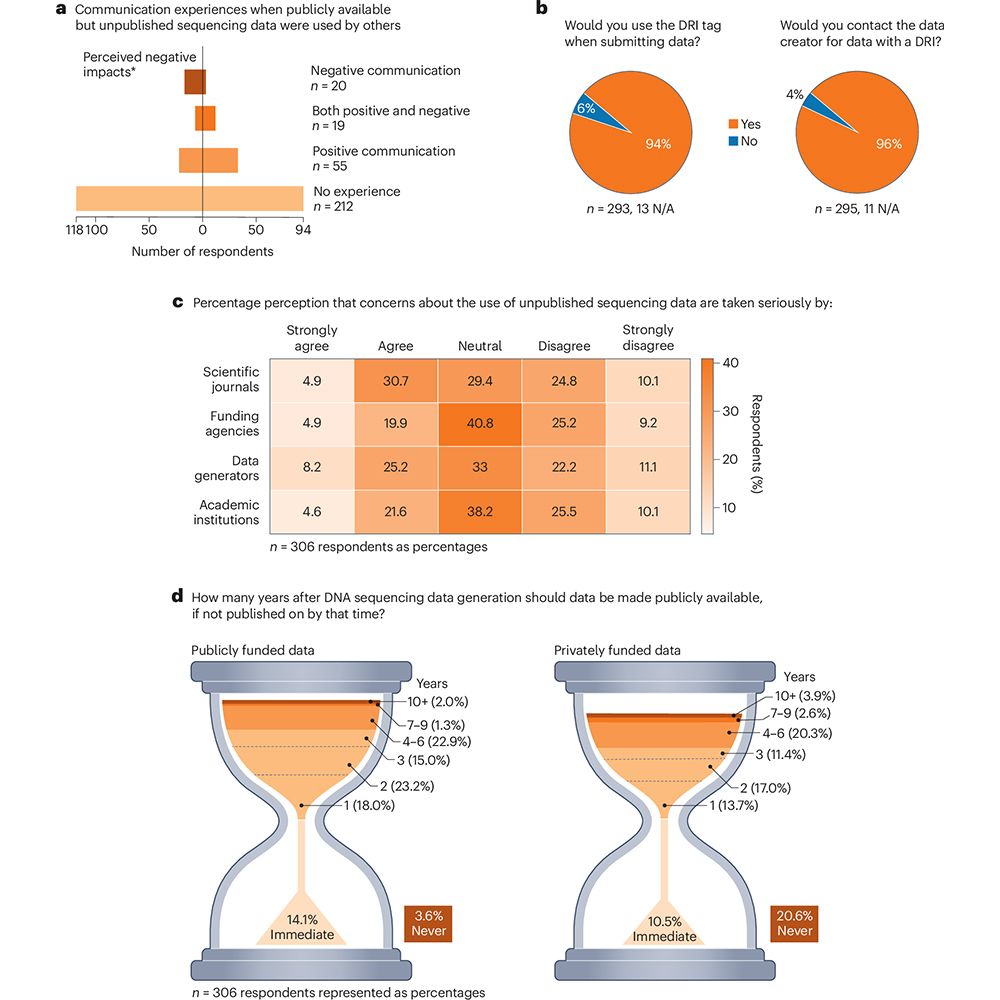
Read all about it in our preprint by👉 doi.org/10.1101/2025...
#Proteomics #AI #MachineLearning

Read all about it in our preprint by👉 doi.org/10.1101/2025...
#Proteomics #AI #MachineLearning
www.nature.com/articles/s41...

www.nature.com/articles/s41...
www.nature.com/articles/s41...
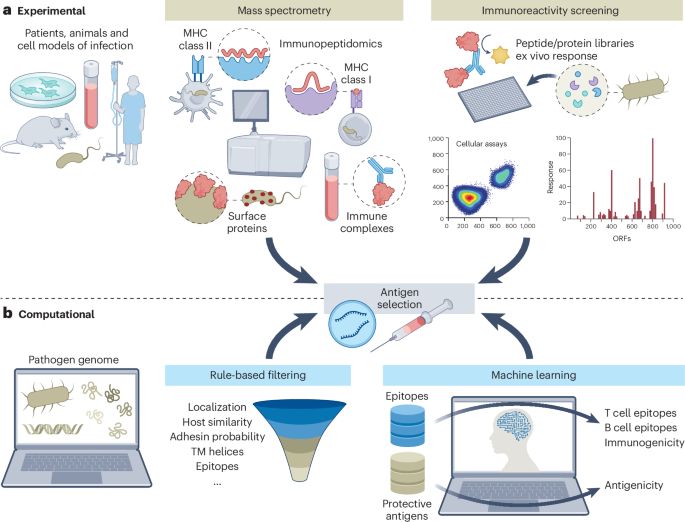
www.nature.com/articles/s41...

