The national programme of Recognition & Rewards is looking for Assistant Professors for an informal dialogue session about content-based career development on 3 March 2026. (1/2)
rrview.nl/calendar_e...

The national programme of Recognition & Rewards is looking for Assistant Professors for an informal dialogue session about content-based career development on 3 March 2026. (1/2)
rrview.nl/calendar_e...

The national programme of Recognition & Rewards is looking for Assistant Professors for an informal dialogue session about content-based career development on 3 March 2026. (1/2)
rrview.nl/calendar_e...
fd24.formdesk.com/universiteit...
#AssistantProfessor #Netherlands #Networking #Lobby
CC: @unileiden.bsky.social
If you are an Assistant professor at @unileiden.bsky.social or LUMC, consider joining this meeting to build a string network.
Sign up: fd24.formdesk.com/universiteit...

fd24.formdesk.com/universiteit...
#AssistantProfessor #Netherlands #Networking #Lobby
CC: @unileiden.bsky.social
If you are an Assistant professor at @unileiden.bsky.social or LUMC, consider joining this meeting to build a string network.
Sign up: fd24.formdesk.com/universiteit...

If you are an Assistant professor at @unileiden.bsky.social or LUMC, consider joining this meeting to build a string network.
Sign up: fd24.formdesk.com/universiteit...
We are working on an exciting additional program. Stay tuned!
Mailing list: fd24.formdesk.com/universiteit...
#ChemSky #ChemBio #ChemBioTalks
We are working on an exciting additional program. Stay tuned!
Mailing list: fd24.formdesk.com/universiteit...
#ChemSky #ChemBio #ChemBioTalks
If you are an Assistant Professor or equivalent in the Netherlands, we invite you to join APNet, a vibrant community created by and for Assistant Professors across the Netherlands. (1/4)
ap-net.nl/register/


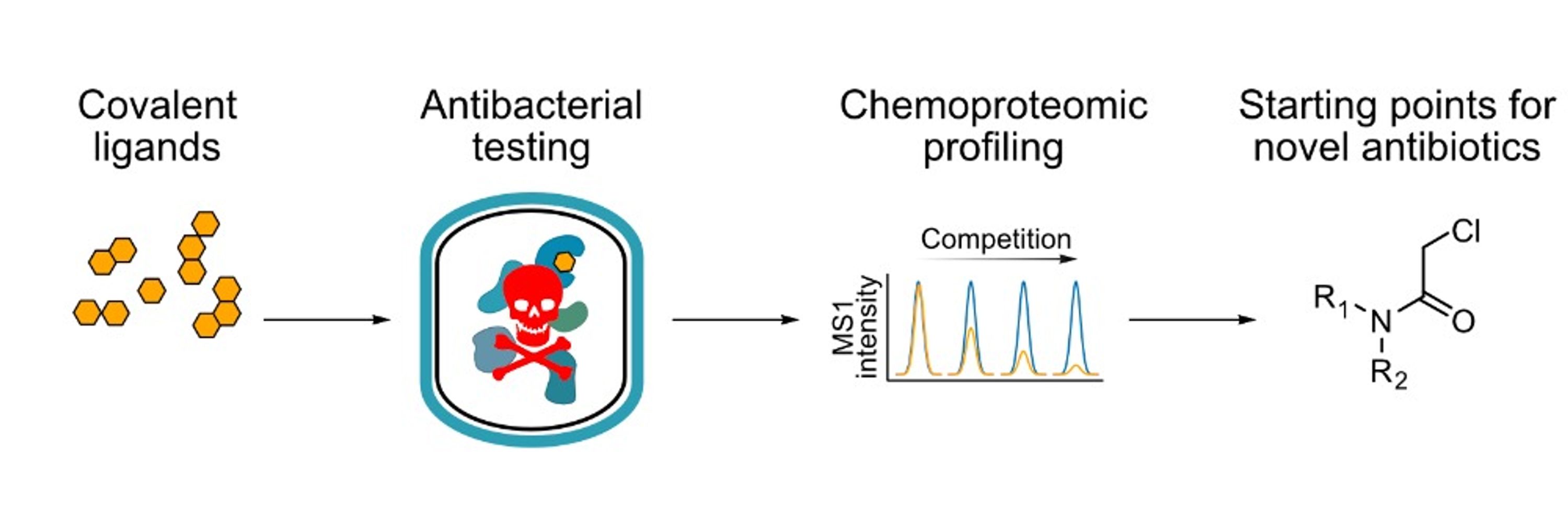
If you are interested in #ChemicalProteomics and/or targeted protein degradation #TPD, you will not want to miss it.
#ChemSky #ChemBio #DrugDiscovery #degrader
If you are interested, make sure to come by!
www.universiteitleiden.nl/en/science/l...
#ChemBio #ChemSky #ChemicalProteomics #TPD

If you are interested in #ChemicalProteomics and/or targeted protein degradation #TPD, you will not want to miss it.
#ChemSky #ChemBio #DrugDiscovery #degrader
If you are interested, make sure to come by!
www.universiteitleiden.nl/en/science/l...
#ChemBio #ChemSky #ChemicalProteomics #TPD

If you are interested, make sure to come by!
www.universiteitleiden.nl/en/science/l...
#ChemBio #ChemSky #ChemicalProteomics #TPD
#NaturalProducts

#NaturalProducts
www.nature.com/articles/s41...
#Plants #NaturalProducts


www.nature.com/articles/s41...
#Plants #NaturalProducts
#Plants #NaturalProducts #GeneClusters #DrugDiscovery #ChemBio #ChemSky #Food #Health

#Plants #NaturalProducts #GeneClusters #DrugDiscovery #ChemBio #ChemSky #Food #Health
Check out our #Biochemistry2026 meeting in Würzburg from March 16th to 18th.
Deadlines for reduced early bird registration fee and abstracts have been extended to January 28th.
veranstaltungen.gdch.de/microsite/in...

Check out our #Biochemistry2026 meeting in Würzburg from March 16th to 18th.
Deadlines for reduced early bird registration fee and abstracts have been extended to January 28th.
veranstaltungen.gdch.de/microsite/in...
The @knaw-nl.bsky.social may have the right funding for you:
- you have defended your PhD 3 to 7 years ago
- you work in academia in the Netherlands
- up to 10,000 €
- deadline is February 2nd
www.knaw.nl/en/funds-and...
The @knaw-nl.bsky.social may have the right funding for you:
- you have defended your PhD 3 to 7 years ago
- you work in academia in the Netherlands
- up to 10,000 €
- deadline is February 2nd
www.knaw.nl/en/funds-and...
CC: @leidenscience.bsky.social
We have a great program for our LED3 Lectures for the remaining academic year. Check out the program and mark your calendars!
www.universiteitleiden.nl/en/science/l...
#DrugDiscovery #ChemSky #ChemBio

CC: @leidenscience.bsky.social
If you are interested, make sure to come by!
www.universiteitleiden.nl/en/science/l...
#ChemBio #ChemSky #Plants #Metabolites

If you are interested, make sure to come by!
www.universiteitleiden.nl/en/science/l...
#ChemBio #ChemSky #Plants #Metabolites
en.gdch.de/network-s...

en.gdch.de/network-s...
Jennifer Andexer, Helge Bode, Stephan Hammer, Akane Kawamura, Alexandria (Ali) Deliz Liang, Carina de Oliveira Mann, Stefan Raunser, Schraga Schwartz, Pierre Stallforth, Carine Tisné, Satpal Virdee, Wilfred van der Donk
#Chemistry #Meeting
CC: @gdch.de
Jennifer Andexer, Helge Bode, Stephan Hammer, Akane Kawamura, Alexandria (Ali) Deliz Liang, Carina de Oliveira Mann, Stefan Raunser, Schraga Schwartz, Pierre Stallforth, Carine Tisné, Satpal Virdee, Wilfred van der Donk
#Chemistry #Meeting
CC: @gdch.de
Apply for our Meeting Award to get:
✅ 500 € grant for the #Biochemistry2026 conference
✅ 1 year of free Fachgruppe Biochemie membership
Apply now: en.gdch.de/network-s...
Share with a #ECR who needs to see this!
#Chemistry #ChemBio #PhD @gdch.de

Apply for our Meeting Award to get:
✅ 500 € grant for the #Biochemistry2026 conference
✅ 1 year of free Fachgruppe Biochemie membership
Apply now: en.gdch.de/network-s...
Share with a #ECR who needs to see this!
#Chemistry #ChemBio #PhD @gdch.de
Registration is now open and you can submit an abstract until January 10th, 2026: veranstaltungen.gdch...
(1/2)

Registration is now open and you can submit an abstract until January 10th, 2026: veranstaltungen.gdch...
(1/2)
Registration is now open and you can submit an abstract until January 10th, 2026: veranstaltungen.gdch...
(1/2)




Great list @stephanhacker2.bsky.social and thanks for sharing.
Great list @stephanhacker2.bsky.social and thanks for sharing.
stay tuned!!
www.nature.com/articles/s41...

stay tuned!!

