Koina aims to transform how #proteomics uses machine learning. You no longer need to be a tech wizard to use ML and now can easily run #ML models. Integrated with FragPipe, Skyline and EncyclopeDIA!
www.nature.com/articles/s41...

Koina aims to transform how #proteomics uses machine learning. You no longer need to be a tech wizard to use ML and now can easily run #ML models. Integrated with FragPipe, Skyline and EncyclopeDIA!
www.nature.com/articles/s41...
Huge thanks to everyone, and special thanks to @stephanhacker2.bsky.social and @pzanon.bsky.social
Our study on "Profiling the proteome-wide selectivity of diverse electrophiles" is published in Nature Chemistry.(1/7)
www.nature.com/articles/s41...

Huge thanks to everyone, and special thanks to @stephanhacker2.bsky.social and @pzanon.bsky.social
Our study on "Profiling the proteome-wide selectivity of diverse electrophiles" is published in Nature Chemistry.(1/7)
www.nature.com/articles/s41...

Our study on "Profiling the proteome-wide selectivity of diverse electrophiles" is published in Nature Chemistry.(1/7)
www.nature.com/articles/s41...
Presenters: Eduard Sabidó and Brendan MacLean
When: Tuesday, September 16, 8am (Pacific Time)
Register Now ... skyline.ms/project/home...
#massspec #proteomics
Presenters: Eduard Sabidó and Brendan MacLean
When: Tuesday, September 16, 8am (Pacific Time)
Register Now ... skyline.ms/project/home...
#massspec #proteomics



The proteomics data for our study on this headed by @pzanon.bsky.social are now public on @pride-ebi.bsky.social: www.ebi.ac.uk/pride/archiv...
Full story: chemrxiv.org/engage/chemr...
#ChemBio #ChemSky #ChemicalProteomics

The proteomics data for our study on this headed by @pzanon.bsky.social are now public on @pride-ebi.bsky.social: www.ebi.ac.uk/pride/archiv...
Full story: chemrxiv.org/engage/chemr...
#ChemBio #ChemSky #ChemicalProteomics
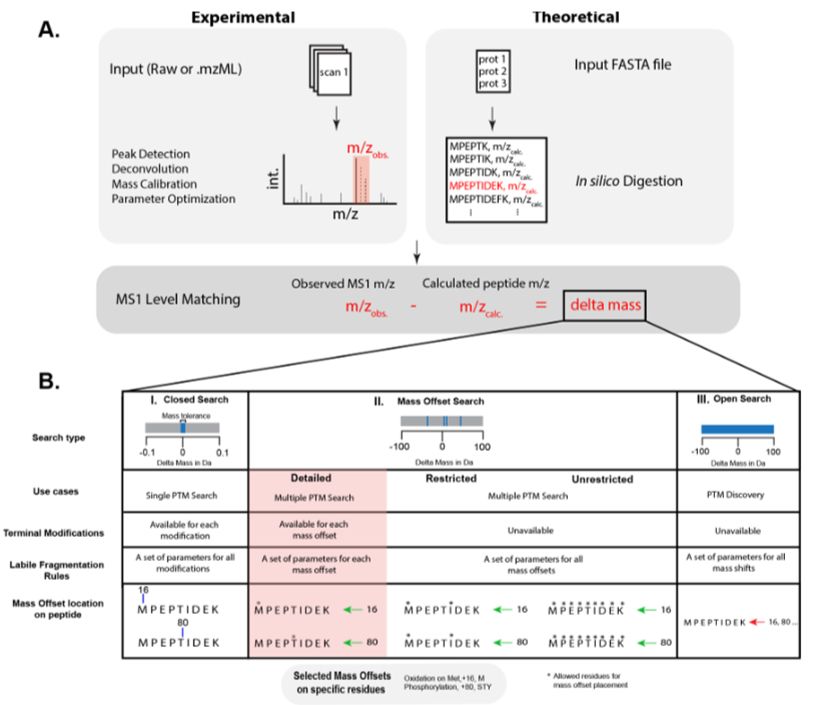



We use zero-distance⚡photo-crosslinking⚡to reveal direct protein-DNA interactions in living cells, enabling quantitative analysis of the DNA-interacting proteome on a timescale of minutes. #DNA #Chromatin #Proteomics
www.cell.com/cell/fulltex...
Sciex is no longer providing funding for Skyline/Proteowizard as part of the multi-vendor agreement to keep Skyline supported across all major vendors.
I can see the arguments from both sides, just a shame it's resulted in this.


www.nature.com/articles/s41...
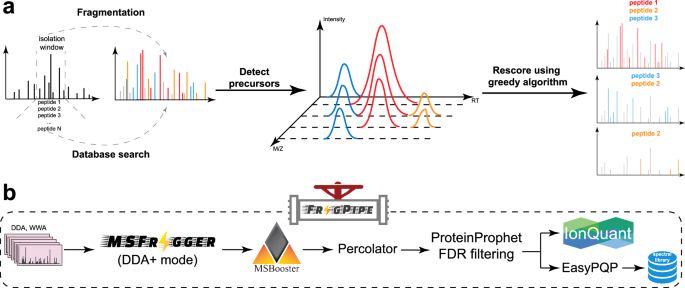
www.nature.com/articles/s41...

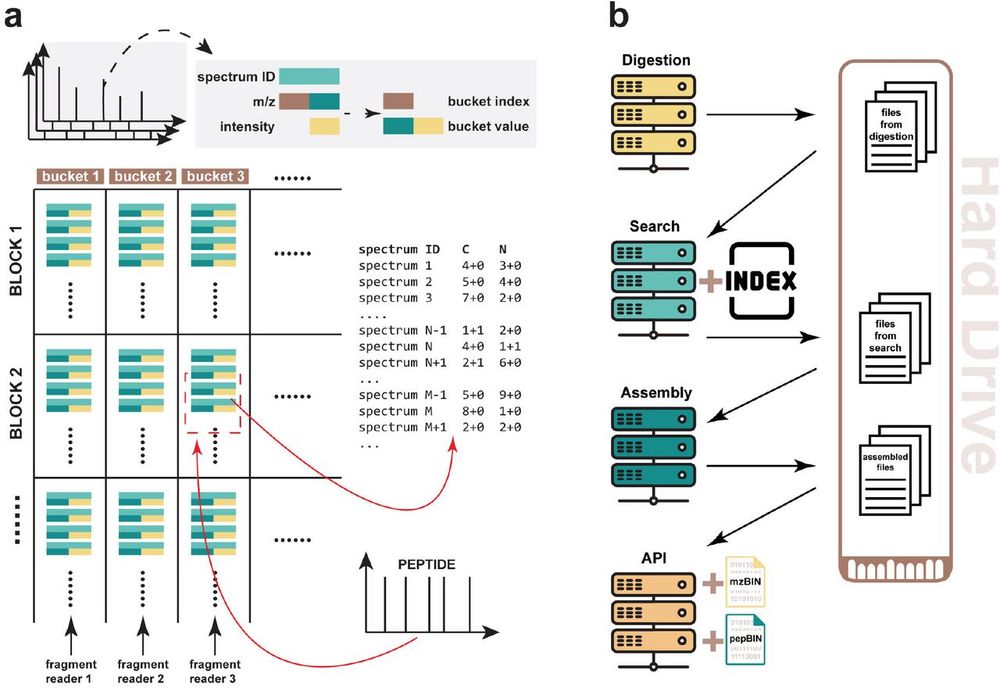

---
#proteomics #prot-paper

---
#proteomics #prot-paper



