
Qian Cheng
@qchengx1e3.bsky.social
Senior Editor at Nature Communications (Computational Biology, AI for Biotechnology). Science communications. PhD (Cantab). Views my own.
https://www.nature.com/subjects/computational-biology-and-bioinformatics/ncomms
https://www.nature.com/subjects/computational-biology-and-bioinformatics/ncomms
Benchmarking scHi-C embedding methods shows that data representation, preprocessing options, and biological settings play a more important role in identifying cell states. @fulaijin.bsky.social
www.nature.com/articles/s41...
www.nature.com/articles/s41...

A comprehensive benchmark of single-cell Hi-C embedding tools - Nature Communications
Embedding is a key step in single-cell Hi-C analysis to identify cell states. Here, the authors benchmark 13 embedding methods in 10 scHi-C datasets. They find that data representation, preprocessing ...
www.nature.com
October 14, 2025 at 5:52 PM
Benchmarking scHi-C embedding methods shows that data representation, preprocessing options, and biological settings play a more important role in identifying cell states. @fulaijin.bsky.social
www.nature.com/articles/s41...
www.nature.com/articles/s41...
A biophysically principled sequence model IceQream characterises complex Transcription Factor-DNA interactions from chromosome accessibility analysis.
@Akhiad.bsky.social
#BiotechNatureComms
www.nature.com/articles/s41...
@Akhiad.bsky.social
#BiotechNatureComms
www.nature.com/articles/s41...

IceQream: Quantitative chromosome accessibility analysis using physical TF models - Nature Communications
Cis regulatory elements endow genomes with sequence-encoded logic to drive cellular differentiation. Here, the authors introduce a biophysically principled sequence model that characterises complex TF...
www.nature.com
October 14, 2025 at 5:52 PM
A biophysically principled sequence model IceQream characterises complex Transcription Factor-DNA interactions from chromosome accessibility analysis.
@Akhiad.bsky.social
#BiotechNatureComms
www.nature.com/articles/s41...
@Akhiad.bsky.social
#BiotechNatureComms
www.nature.com/articles/s41...
A multi-scale, multi-context, and interpretable mapping strategy to map cells across space, time, and disease. @pcnmartin.bsky.social @wonkj.bsky.social
#spatialMapping #spatialOmics #BiotechNatureComms
www.nature.com/articles/s41...
#spatialMapping #spatialOmics #BiotechNatureComms
www.nature.com/articles/s41...

Multi-scale and multi-context interpretable mapping of cell states across heterogeneous spatial samples - Nature Communications
The alignment of heterogeneous spatial samples has become a growing challenge. Here, authors present a multi-scale, multi-context, and interpretable mapping strategy to map cells across space, time, a...
www.nature.com
August 22, 2025 at 2:18 PM
A multi-scale, multi-context, and interpretable mapping strategy to map cells across space, time, and disease. @pcnmartin.bsky.social @wonkj.bsky.social
#spatialMapping #spatialOmics #BiotechNatureComms
www.nature.com/articles/s41...
#spatialMapping #spatialOmics #BiotechNatureComms
www.nature.com/articles/s41...
Using microfluidics and endogenous reporters, single cells are tracked to reveal how temporal dosing rewires chromatin in a model with near single-cell accuracy. @stevenwsmeal.bsky.social @robineclee.bsky.social #BiotechNatureComms
www.nature.com/articles/s41...
www.nature.com/articles/s41...

Time-varying stimuli that prolong IKK activation promote nuclear remodeling and mechanistic switching of NF-κB dynamics - Nature Communications
Cells rely on limited numbers of transmembrane receptors to process signals from dynamic microenvironments. Using microfluidics and endogenous reporters, the authors track single cells to reveal how t...
www.nature.com
August 14, 2025 at 8:51 PM
Using microfluidics and endogenous reporters, single cells are tracked to reveal how temporal dosing rewires chromatin in a model with near single-cell accuracy. @stevenwsmeal.bsky.social @robineclee.bsky.social #BiotechNatureComms
www.nature.com/articles/s41...
www.nature.com/articles/s41...
A computational framework for single cell and spatial alternative splicing analysis with Nanopore long read sequencing
@nancyrzhang.bsky.social
#BiotechNatureComms
www.nature.com/articles/s41...
@nancyrzhang.bsky.social
#BiotechNatureComms
www.nature.com/articles/s41...
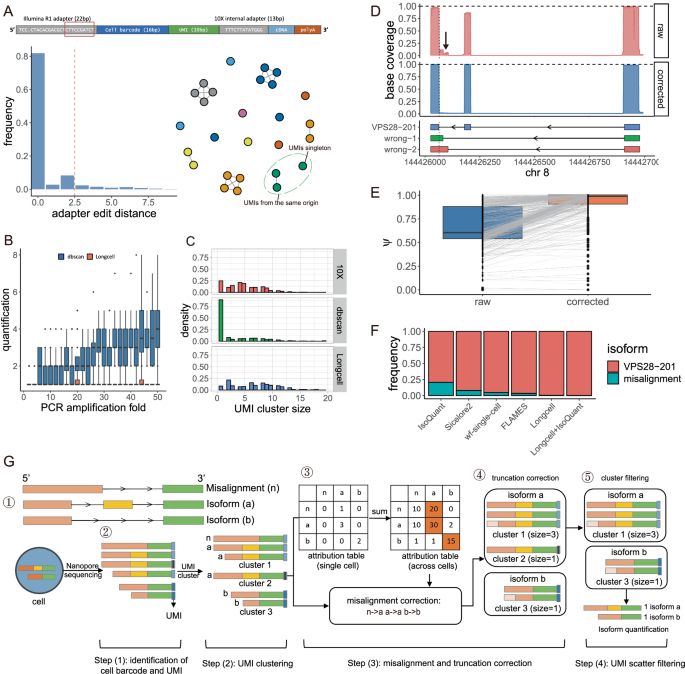
Single cell and spatial alternative splicing analysis with Nanopore long read sequencing - Nature Communications
Single-cell isoform quantification using Nanopore long reads remains limited by sequencing errors. Here, authors present Longcell, a computational framework that corrects these errors and uncovers spl...
www.nature.com
July 22, 2025 at 1:28 PM
A computational framework for single cell and spatial alternative splicing analysis with Nanopore long read sequencing
@nancyrzhang.bsky.social
#BiotechNatureComms
www.nature.com/articles/s41...
@nancyrzhang.bsky.social
#BiotechNatureComms
www.nature.com/articles/s41...
A computational tool #SCALPEL for #isoform quantification at the single-cell level using 3’ scRNA-seq data @franzake.bsky.social @mireyaplass.bsky.social #BiotechNatureComms
doi.org/10.1038/s414...
doi.org/10.1038/s414...
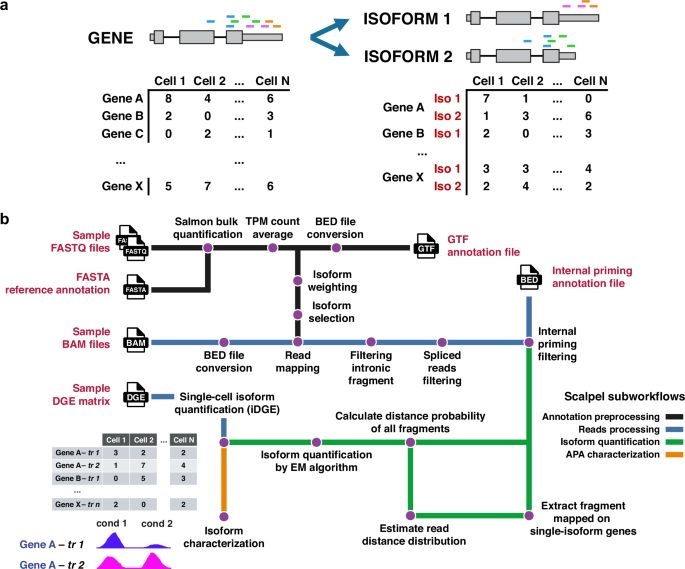
Quantification of transcript isoforms at the single-cell level using SCALPEL - Nature Communications
Single-cell RNA-seq facilitates the study of transcriptome diversity in individual cells. Here, authors introduce a tool for isoform quantification at the single-cell level using 3’ scRNA-seq data, co...
doi.org
July 13, 2025 at 5:41 PM
A computational tool #SCALPEL for #isoform quantification at the single-cell level using 3’ scRNA-seq data @franzake.bsky.social @mireyaplass.bsky.social #BiotechNatureComms
doi.org/10.1038/s414...
doi.org/10.1038/s414...
A fully automated AI ImageJ plugin that detects and classifies #exocytosis events from synaptic transmission to single-vesicle fusion. @alishaib.bsky.social #BiotechNatureComms
doi.org/10.1038/s414...
doi.org/10.1038/s414...
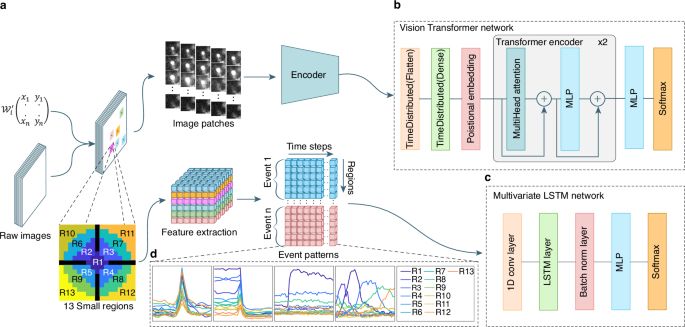
Highly adaptable deep-learning platform for automated detection and analysis of vesicle exocytosis - Nature Communications
Activity recognition in live-cell imaging is laborious. Here, authors present, IVEA, a fully automated AI ImageJ plugin, that efficiently detects and classifies exocytosis events, from synaptic transm...
doi.org
July 13, 2025 at 5:39 PM
A fully automated AI ImageJ plugin that detects and classifies #exocytosis events from synaptic transmission to single-vesicle fusion. @alishaib.bsky.social #BiotechNatureComms
doi.org/10.1038/s414...
doi.org/10.1038/s414...
ToxACoL is an endpoint-aware and task-focused compound representation learning paradigm for #Acute_Toxicity_Assessment #BiotechNatureComms
doi.org/10.1038/s414...
doi.org/10.1038/s414...
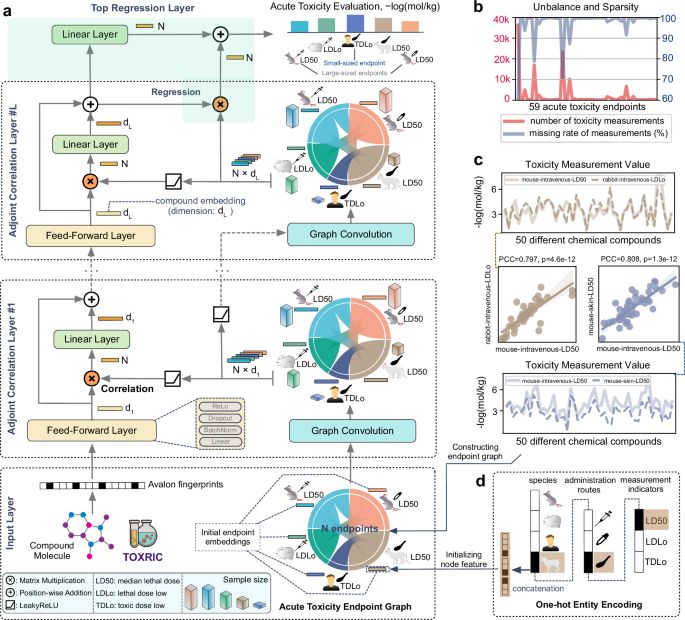
ToxACoL: an endpoint-aware and task-focused compound representation learning paradigm for acute toxicity assessment - Nature Communications
Multi-species acute systemic toxicity assessment is fundamental for chemical classification, labelling and risk management. Here, the authors propose a machine learning paradigm, Adjoint Correlation L...
doi.org
July 7, 2025 at 3:16 PM
ToxACoL is an endpoint-aware and task-focused compound representation learning paradigm for #Acute_Toxicity_Assessment #BiotechNatureComms
doi.org/10.1038/s414...
doi.org/10.1038/s414...
Binarized gene expression data can be concatenated with chromatin accessibility data for effective and integrated cell clustering @zhengdy.bsky.social @einsteinmededu.bsky.social #BiotechNatureComms
doi.org/10.1038/s414...
doi.org/10.1038/s414...
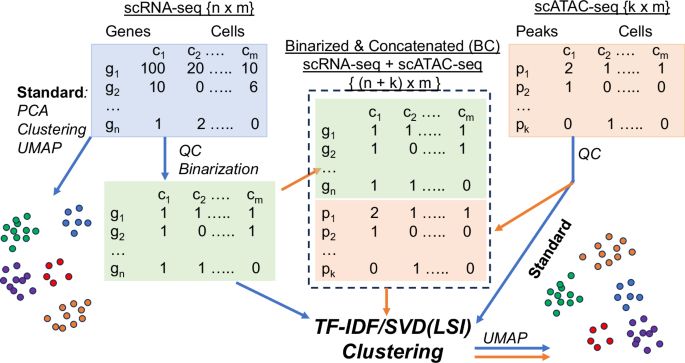
Facilitate integrated analysis of single cell multiomic data by binarizing gene expression values - Nature Communications
Individual data types in a single cell multi-omic assay are often processed and clustered independently before combined for further analysis. Here, authors showed that binarized gene expression d...
doi.org
July 7, 2025 at 3:15 PM
Binarized gene expression data can be concatenated with chromatin accessibility data for effective and integrated cell clustering @zhengdy.bsky.social @einsteinmededu.bsky.social #BiotechNatureComms
doi.org/10.1038/s414...
doi.org/10.1038/s414...
GPerturb detects and quantifies gene-level effects of perturbations, with uncertainty estimates, revealing complex biological interactions. @cwcyau.bsky.social
doi.org/10.1038/s414...
doi.org/10.1038/s414...
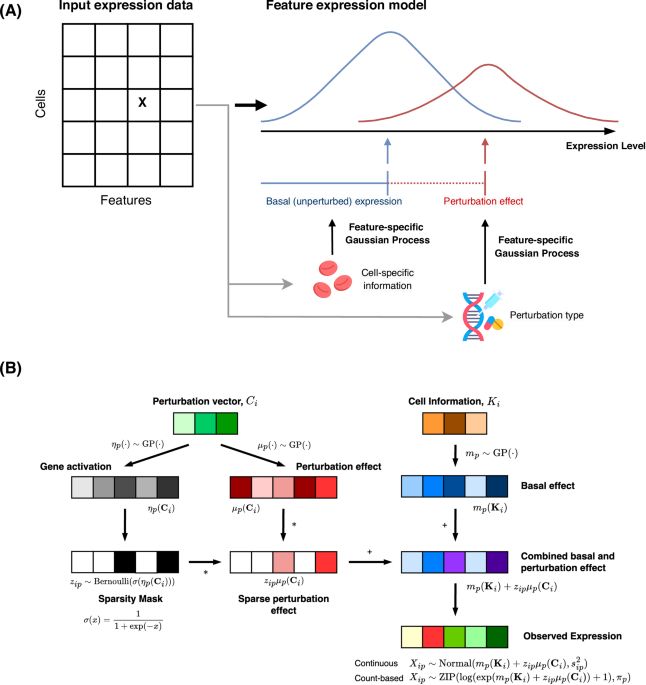
GPerturb: Gaussian process modelling of single-cell perturbation data - Nature Communications
Single-cell CRISPR screens enable high-throughput analysis of how genetic changes affect individual cells. Here, authors present GPerturb, a method that accurately detects and quantifies gene-level ef...
doi.org
July 7, 2025 at 3:13 PM
GPerturb detects and quantifies gene-level effects of perturbations, with uncertainty estimates, revealing complex biological interactions. @cwcyau.bsky.social
doi.org/10.1038/s414...
doi.org/10.1038/s414...
A virtual single-cell NFκB signalling networks that recapitulate heterogenous experimental stimulus-response dynamics @xiaoluguo.bsky.social #BiotechNatureComms
doi.org/10.1038/s414...
doi.org/10.1038/s414...
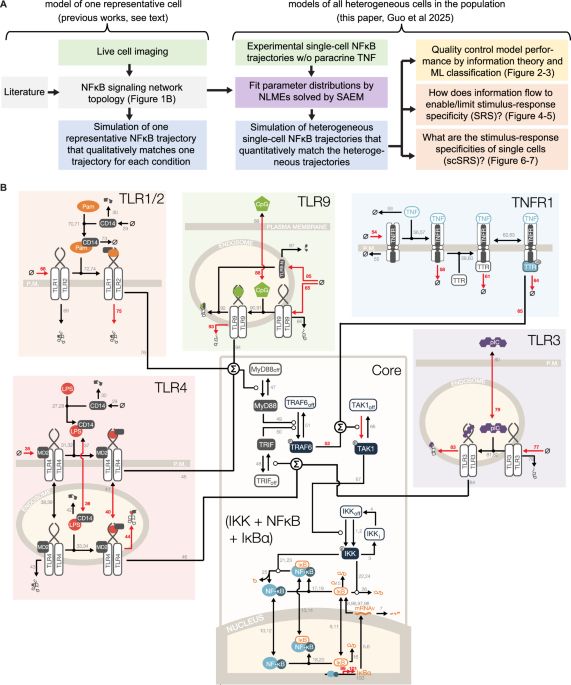
Modeling heterogeneous signaling dynamics of macrophages reveals principles of information transmission in stimulus responses - Nature Communications
To develop virtual cells, mathematical models must account for the heterogeneity of single cells. Here, authors develop virtual single cell NFκB signalling networks that recapitulate heterogenous expe...
doi.org
July 7, 2025 at 3:13 PM
A virtual single-cell NFκB signalling networks that recapitulate heterogenous experimental stimulus-response dynamics @xiaoluguo.bsky.social #BiotechNatureComms
doi.org/10.1038/s414...
doi.org/10.1038/s414...
DOLPHIN: a deep learning method that enables exon- and junction-level analysis to improve cell representation and detect alternative #splicing. Ding Lab junding.lab.mcgill.ca #BiotechNatureComms
doi.org/10.1038/s414...
doi.org/10.1038/s414...
Ding Lab
website description
junding.lab.mcgill.ca
July 7, 2025 at 3:09 PM
DOLPHIN: a deep learning method that enables exon- and junction-level analysis to improve cell representation and detect alternative #splicing. Ding Lab junding.lab.mcgill.ca #BiotechNatureComms
doi.org/10.1038/s414...
doi.org/10.1038/s414...
A deep learning and immune-fluorescence-guided cell labelling method for accurate cell pattern identification. #BiotechNatureComms
doi.org/10.1038/s414...
doi.org/10.1038/s414...
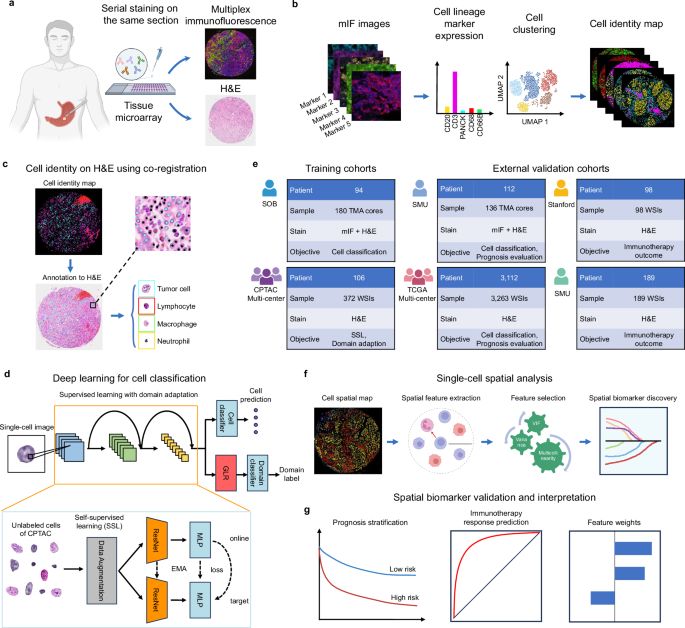
Automated cell annotation and classification on histopathology for spatial biomarker discovery - Nature Communications
Histopathology is essential for diagnosing cancer, but cell annotation remains unreliable and inefficient. Here, authors develop a method using deep learning and immune-fluorescence-guided cell labell...
doi.org
July 7, 2025 at 3:08 PM
A deep learning and immune-fluorescence-guided cell labelling method for accurate cell pattern identification. #BiotechNatureComms
doi.org/10.1038/s414...
doi.org/10.1038/s414...
Cell Marker Accordion is a user-friendly platform providing robust automatic cell annotation of #single_cell and spatial populations @rdds-lab.bsky.social #BiotechNatureComms
doi.org/10.1038/s414...
doi.org/10.1038/s414...
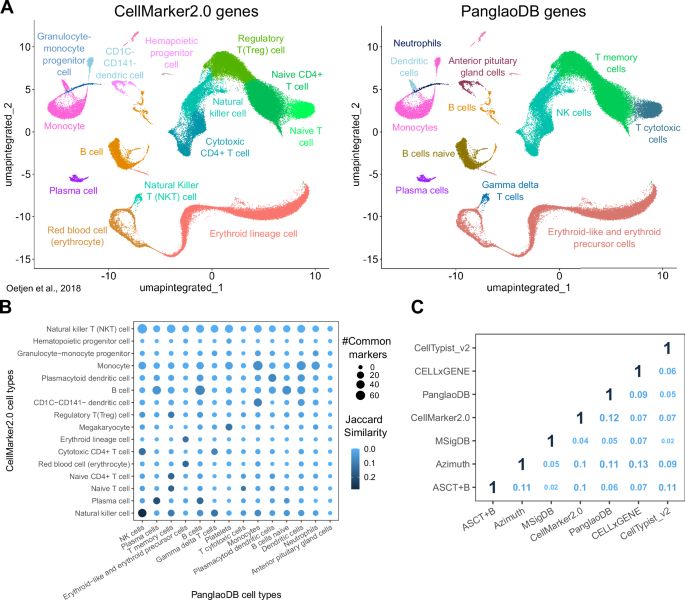
Cell Marker Accordion: interpretable single-cell and spatial omics annotation in health and disease - Nature Communications
Accurate cell type annotation is a major challenge in single-cell and spatial omics. Here, authors present a user-friendly platform providing robust automatic annotation and enhanced biological interp...
doi.org
July 7, 2025 at 3:08 PM
Cell Marker Accordion is a user-friendly platform providing robust automatic cell annotation of #single_cell and spatial populations @rdds-lab.bsky.social #BiotechNatureComms
doi.org/10.1038/s414...
doi.org/10.1038/s414...
A Bayesian optimisation-based iterative experimental design framework that integrates data generation, modelling, and optimisation to accelerate cell culture media development #BiotechNatureComms
doi.org/10.1038/s414...
doi.org/10.1038/s414...
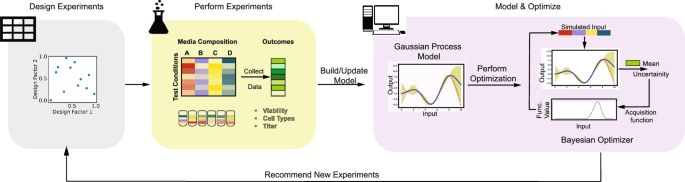
Accelerating cell culture media development using Bayesian optimization-based iterative experimental design - Nature Communications
Optimising operational conditions for biological systems is a complex task. Here, authors present a Bayesian optimisation-based iterative experimental design framework that integrates data generation,...
doi.org
July 7, 2025 at 3:05 PM
A Bayesian optimisation-based iterative experimental design framework that integrates data generation, modelling, and optimisation to accelerate cell culture media development #BiotechNatureComms
doi.org/10.1038/s414...
doi.org/10.1038/s414...
A multiplexed super-resolution imaging protocol studies how nuclear proteins are distributed relative to each other from the micro to the nanoscale. #BiotechNatureComms
doi.org/10.1038/s414...
doi.org/10.1038/s414...
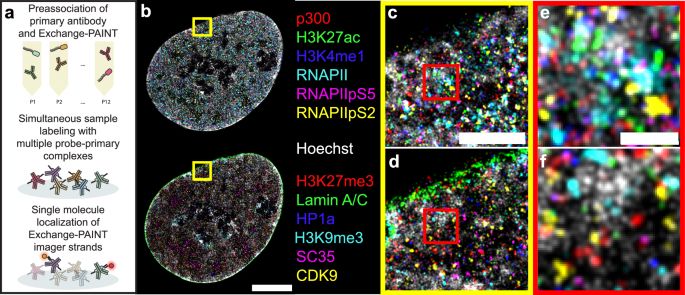
Mapping the nuclear landscape with multiplexed super-resolution fluorescence microscopy - Nature Communications
The cell nucleus coordinates diverse functions. Here, the authors utilise multiplexed super-resolution imaging to study how nuclear proteins are distributed relative to each other from the micro to th...
doi.org
July 7, 2025 at 3:05 PM
A multiplexed super-resolution imaging protocol studies how nuclear proteins are distributed relative to each other from the micro to the nanoscale. #BiotechNatureComms
doi.org/10.1038/s414...
doi.org/10.1038/s414...
Reposted by Qian Cheng
My first task is to hire an Associate/Senior Editor for @natrevelectreng.nature.com. Would you be interested in joining me to shape the journal? Pls DM me if you have any questions💡Apply by 1 Jul.
@natureportfolio.nature.com @springernature.com
springernature.wd3.myworkdayjobs.com/SpringerNatu...
@natureportfolio.nature.com @springernature.com
springernature.wd3.myworkdayjobs.com/SpringerNatu...

Associate or Senior Editor, Nature Reviews Electrical Engineering
Job title: Associate or Senior Editor, Nature Reviews Electrical Engineering Location: Shanghai or New York – hybrid working model Closing date: 1st July 2025 About Springer Nature Group Springer Natu...
springernature.wd3.myworkdayjobs.com
June 13, 2025 at 3:58 PM
My first task is to hire an Associate/Senior Editor for @natrevelectreng.nature.com. Would you be interested in joining me to shape the journal? Pls DM me if you have any questions💡Apply by 1 Jul.
@natureportfolio.nature.com @springernature.com
springernature.wd3.myworkdayjobs.com/SpringerNatu...
@natureportfolio.nature.com @springernature.com
springernature.wd3.myworkdayjobs.com/SpringerNatu...
A versatile information retrieval framework to quantify sample activity and similarity in large-scale profiling data @natcomms.nature.com @alxndrkalinin.bsky.social @shantanu-singh.cc @drannecarpenter.bsky.social
www.nature.com/articles/s41...
www.nature.com/articles/s41...
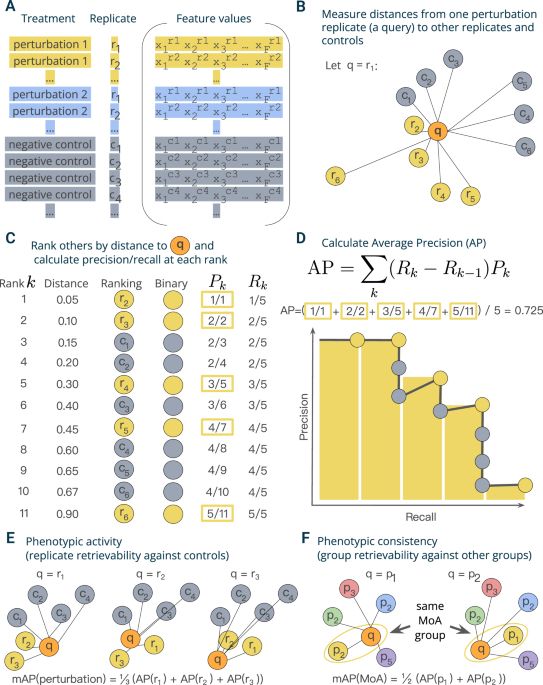
A versatile information retrieval framework for evaluating profile strength and similarity - Nature Communications
Profiling assays measure thousands of features to uncover biological insights but lack reliable methods for quality evaluation. Here, the authors develop a versatile information retrieval framework to...
www.nature.com
June 6, 2025 at 12:26 PM
A versatile information retrieval framework to quantify sample activity and similarity in large-scale profiling data @natcomms.nature.com @alxndrkalinin.bsky.social @shantanu-singh.cc @drannecarpenter.bsky.social
www.nature.com/articles/s41...
www.nature.com/articles/s41...
An AI-assisted fluorescence microscopy system to track mitophagic intermediates in live cells @natcomms.nature.com
www.nature.com/articles/s41...
www.nature.com/articles/s41...
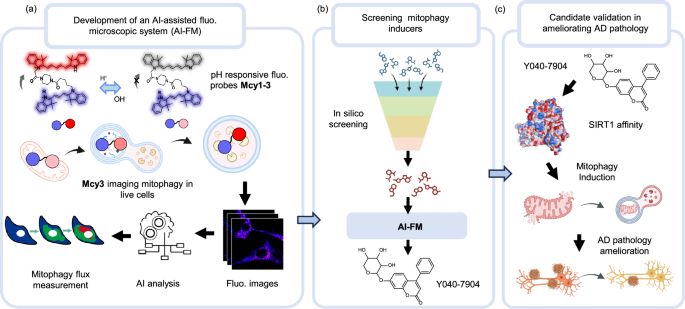
An AI-assisted fluorescence microscopic system for screening mitophagy inducers by simultaneous analysis of mitophagic intermediates - Nature Communications
Mitophagy is crucial for mitochondrial function and is implicated in Alzheimer’s disease. Here, the authors introduce an AI-assisted fluorescence microscopy system to track mitophagic intermediates in...
www.nature.com
June 6, 2025 at 12:25 PM
An AI-assisted fluorescence microscopy system to track mitophagic intermediates in live cells @natcomms.nature.com
www.nature.com/articles/s41...
www.nature.com/articles/s41...
CellFM: A 800-million-parameter foundation model pre-trained on transcriptomics of 100 million human cells. #BiotechNatureComms @natcomms.nature.com
www.nature.com/articles/s41...
www.nature.com/articles/s41...
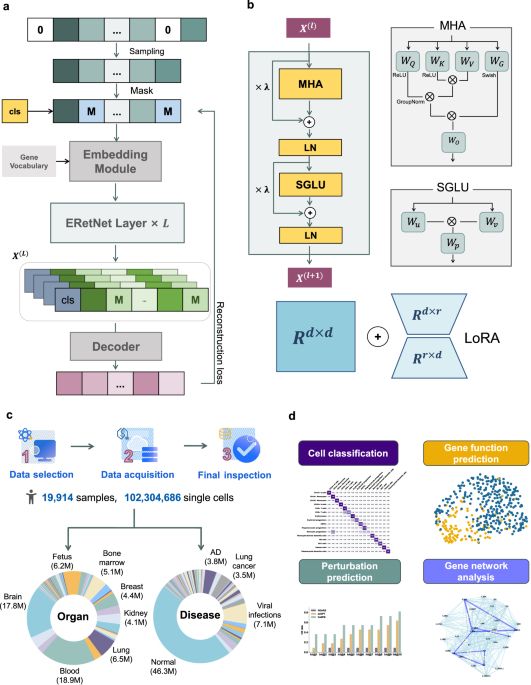
CellFM: a large-scale foundation model pre-trained on transcriptomics of 100 million human cells - Nature Communications
Single-cell sequencing reveals cellular heterogeneity but is challenged by technical noise and batch effects. Here, authors present CellFM, an 800-million-parameter foundation model trained on 100 mil...
www.nature.com
May 28, 2025 at 2:33 PM
CellFM: A 800-million-parameter foundation model pre-trained on transcriptomics of 100 million human cells. #BiotechNatureComms @natcomms.nature.com
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Kasumi identifies local patterns in tissue patches in spatial omics data @tanevski.bsky.social, @saezlab.bsky.social #BiotechNatureComms
www.nature.com/articles/s41...
www.nature.com/articles/s41...
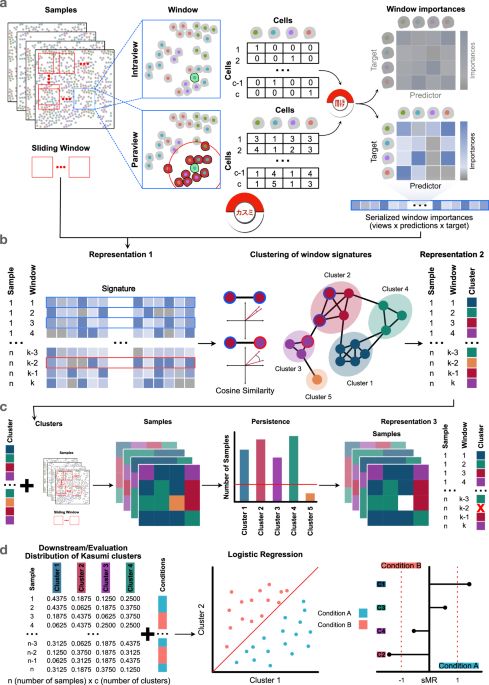
Learning tissue representation by identification of persistent local patterns in spatial omics data - Nature Communications
Spatial omics reveal tissue structures and can aid patient stratification. The authors present a method to identify patterns in tissue patches, enabling analysis of disease progression and treatment r...
www.nature.com
May 8, 2025 at 1:30 PM
Kasumi identifies local patterns in tissue patches in spatial omics data @tanevski.bsky.social, @saezlab.bsky.social #BiotechNatureComms
www.nature.com/articles/s41...
www.nature.com/articles/s41...
A data-efficient, generative AI-based toolkit for designing a diverse set of RNA molecules. #BiotechNatureComms
www.nature.com/articles/s41...
alexgreenlab.org
www.nature.com/articles/s41...
alexgreenlab.org

Green Laboratory
Visit the post for more.
alexgreenlab.org
May 8, 2025 at 1:30 PM
A data-efficient, generative AI-based toolkit for designing a diverse set of RNA molecules. #BiotechNatureComms
www.nature.com/articles/s41...
alexgreenlab.org
www.nature.com/articles/s41...
alexgreenlab.org
EvoWeaver identifies genes working together using evolutionary information and infers protein functions using only genomic sequences. @ahl27.bsky.social @natcomms.nature.com
www.nature.com/articles/s41...
www.nature.com/articles/s41...
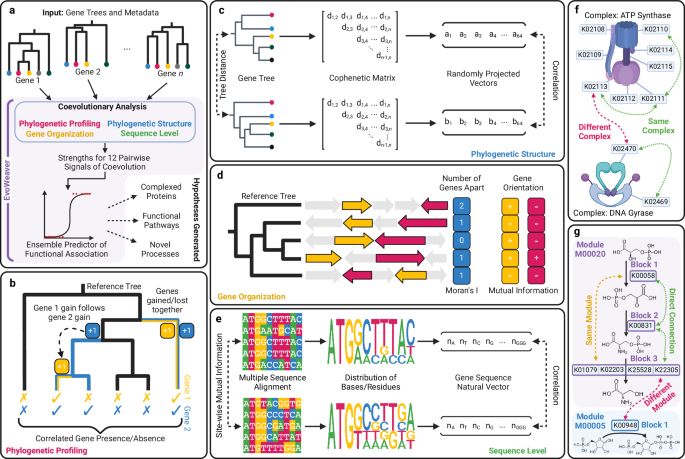
EvoWeaver: large-scale prediction of gene functional associations from coevolutionary signals - Nature Communications
Despite having structures for most proteins, we still do not know their function. Here, authors introduce EvoWeaver, a de novo method to identify genes working together using evolutionary information....
www.nature.com
April 24, 2025 at 5:18 PM
EvoWeaver identifies genes working together using evolutionary information and infers protein functions using only genomic sequences. @ahl27.bsky.social @natcomms.nature.com
www.nature.com/articles/s41...
www.nature.com/articles/s41...
.@jkobject.com @cantinilab.bsky.social @gabrielpeyre.bsky.social present a foundation model trained on 50 million cells, generating a meaningful gene network with zero-shot ability. #AIxBio @natcomms.nature.com
www.nature.com/articles/s41...
www.nature.com/articles/s41...
scPRINT: pre-training on 50 million cells allows robust gene network predictions - Nature Communications
Authors present a state-of-the-art cell foundation model trained on 50 million cells. They show that the model generates a meaningful gene network and has zero-shot ability in various tasks.
www.nature.com
April 21, 2025 at 8:25 PM
.@jkobject.com @cantinilab.bsky.social @gabrielpeyre.bsky.social present a foundation model trained on 50 million cells, generating a meaningful gene network with zero-shot ability. #AIxBio @natcomms.nature.com
www.nature.com/articles/s41...
www.nature.com/articles/s41...
MSFragger-DDA+ is a database search tool that enhances peptide identification by searching the full isolation window. #Proteomics @fcyucn.bsky.social @nesvilab.bsky.social
www.nature.com/articles/s41...
www.nature.com/articles/s41...
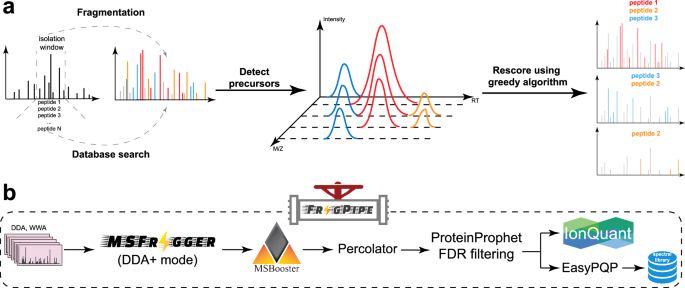
MSFragger-DDA+ enhances peptide identification sensitivity with full isolation window search - Nature Communications
Proteomics often misses co-fragmented peptides in DDA data. Here, the authors introduce MSFragger-DDA+, a database search tool that enhances peptide identification by searching the full isolation wind...
www.nature.com
April 15, 2025 at 4:39 PM
MSFragger-DDA+ is a database search tool that enhances peptide identification by searching the full isolation window. #Proteomics @fcyucn.bsky.social @nesvilab.bsky.social
www.nature.com/articles/s41...
www.nature.com/articles/s41...

