
Donal O’Carroll and coworkers
www.embopress.org/doi/full/10....

Donal O’Carroll and coworkers
www.embopress.org/doi/full/10....
This workshop will teach you transferable skills to support a career as a journal editor or whatever career path you pursue.
Secure Your Spot: www.lifescienceeditors.com/workshop3-si...

This workshop will teach you transferable skills to support a career as a journal editor or whatever career path you pursue.
Secure Your Spot: www.lifescienceeditors.com/workshop3-si...

👉 We discover how sex and development signals are decoded at a single gene locus.
www.nature.com/articles/s41...
👇 Bluetorial

👉 We discover how sex and development signals are decoded at a single gene locus.
www.nature.com/articles/s41...
👇 Bluetorial
Read about the scientific visions of eight dev bio post-docs entering the academic job market
journals.biologists.com/dev/article/...

Read about the scientific visions of eight dev bio post-docs entering the academic job market
journals.biologists.com/dev/article/...


rdcu.be/exXBX

rdcu.be/exXBX
@natbiotech.nature.com: nature.com/articles/s41.... A short tread! (1/n)
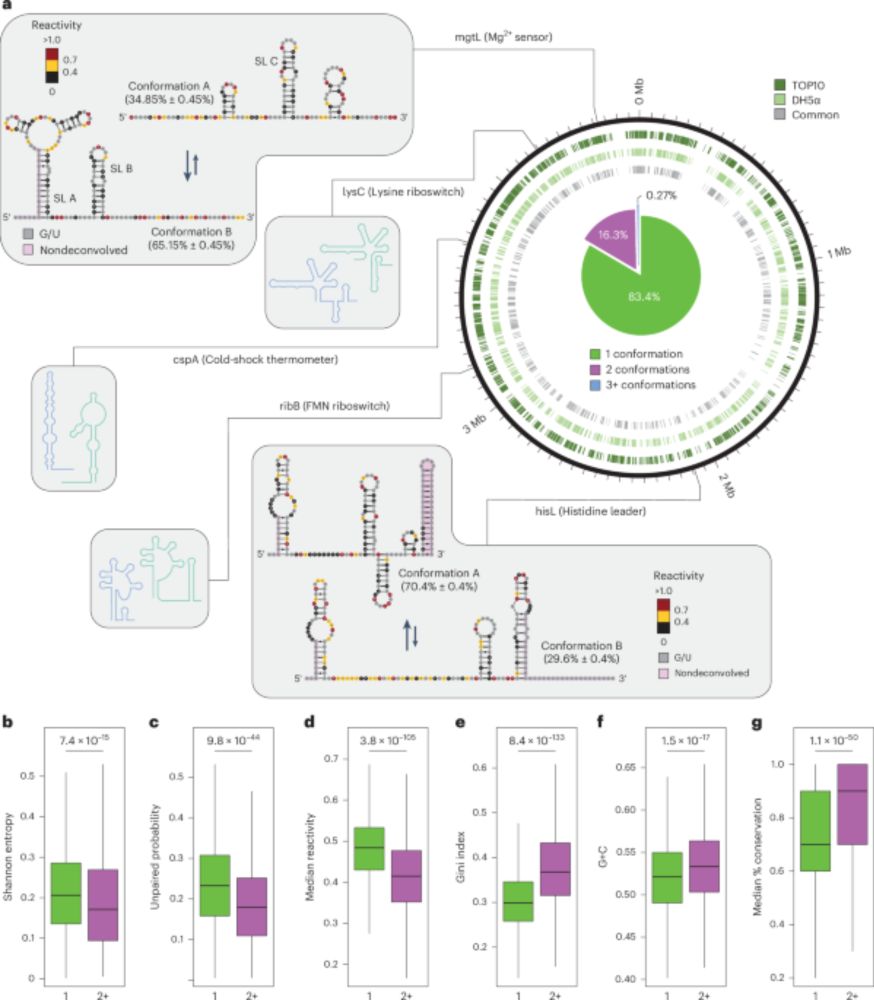
@natbiotech.nature.com: nature.com/articles/s41.... A short tread! (1/n)

Read our interview with study co-author — and UTM Biology prof — Lars Barquist at uoft.me/bI7.

Read our interview with study co-author — and UTM Biology prof — Lars Barquist at uoft.me/bI7.
Check it out here: go.nature.com/4lf6jNk
Check it out here: go.nature.com/4lf6jNk
![3-panel comic. (1) PERSON WITH SHORT HAIR to Person 2: Shining laser pointers at planes is a federal crime. It’s incredibly dangerous. PERSON 2: Oh, because it can blind the pilot? SHORT HAIR: That’s one reason… (2) [Laser pointed at flying airplane] (3) [Cat close to the size of the plane pouncing on airplane and taking it down]](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:cz73r7iyiqn26upot4jtjdhk/bafkreicdr4m3a7s2i232wlvcu7dkwytnrh3tlqfbojtmyg3pzr7a6ibxra@jpeg)
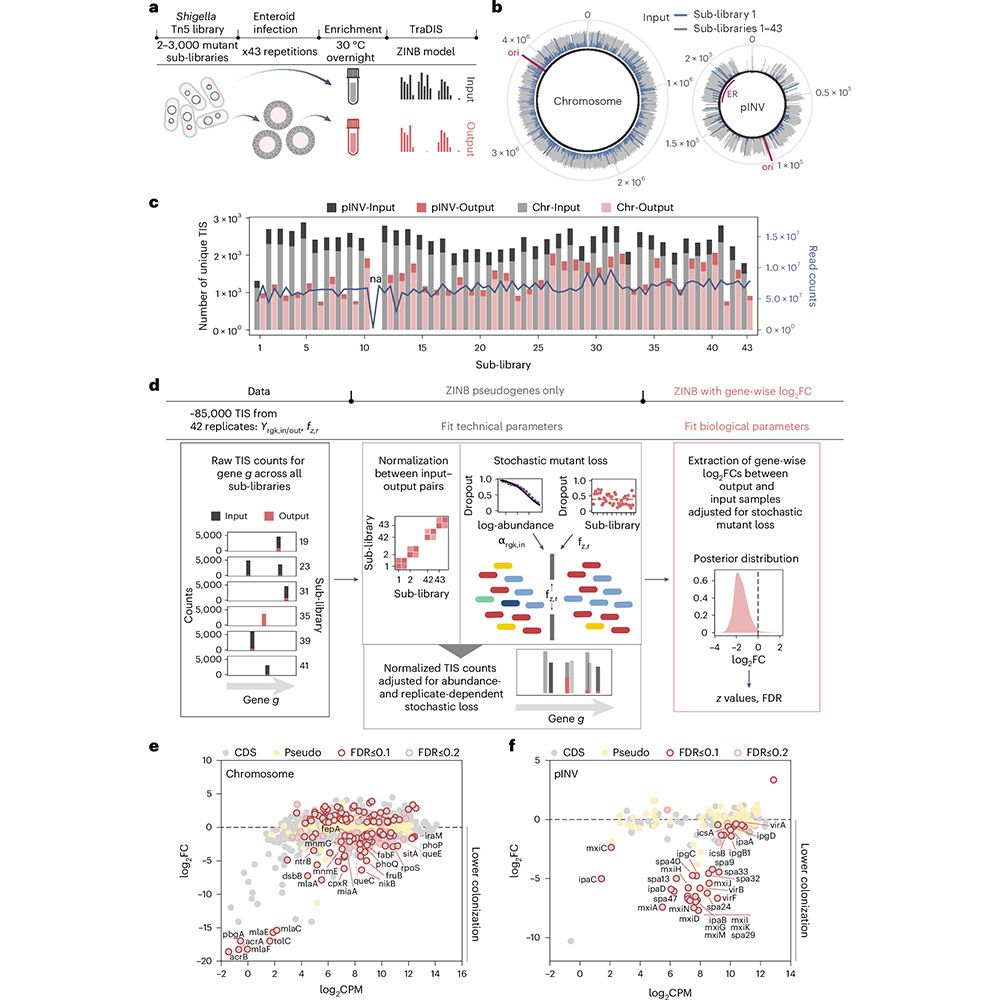
Shoutout to @elisebor.bsky.social, @emmanuel-saliba.bsky.social, @lbarquist.bsky.social, @jorg-vogel-lab.bsky.social, Till Strowig & KC Huang.
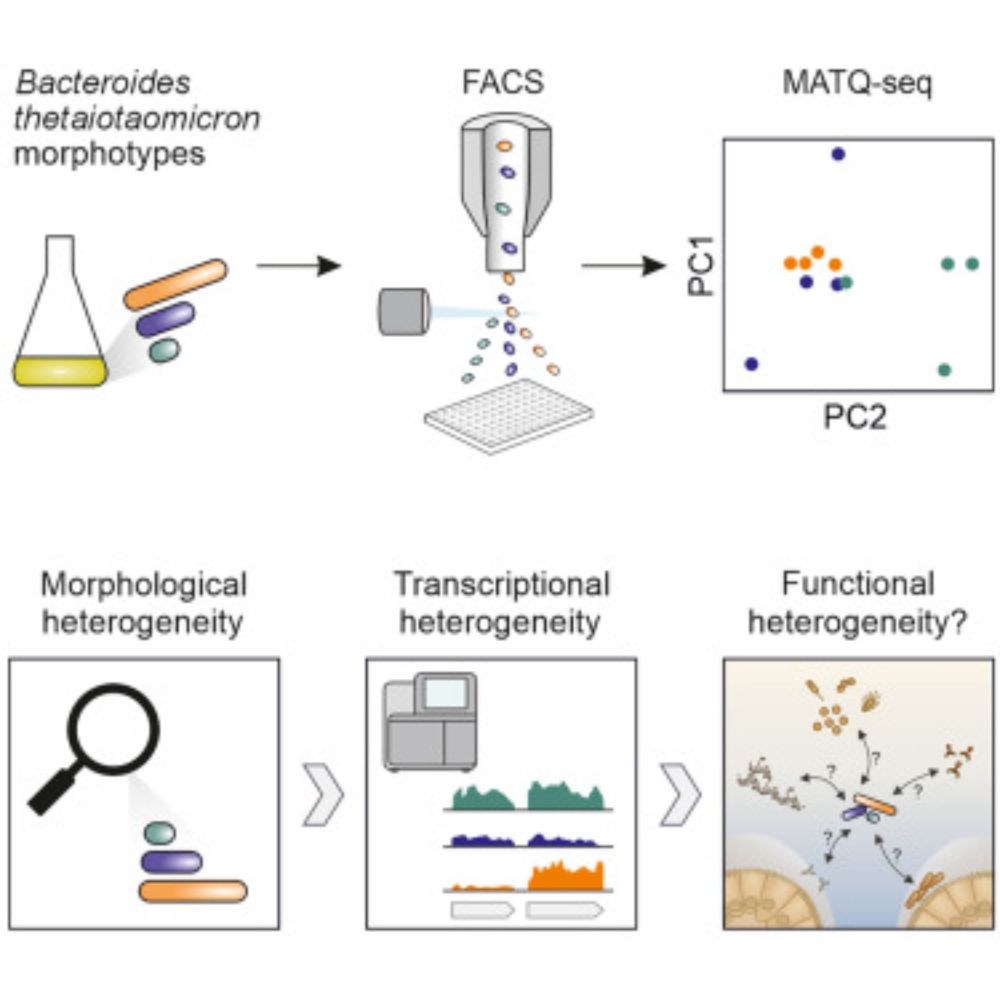
Shoutout to @elisebor.bsky.social, @emmanuel-saliba.bsky.social, @lbarquist.bsky.social, @jorg-vogel-lab.bsky.social, Till Strowig & KC Huang.
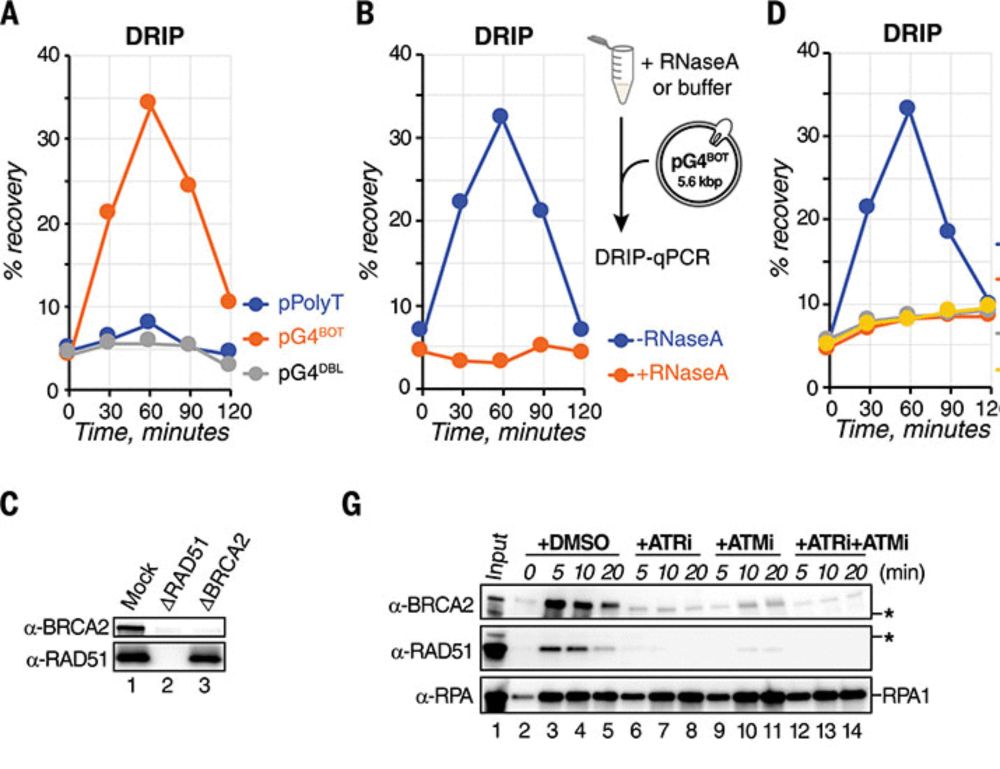
We established mudRapp-seq, a spatial transcriptomic technique that combines direct RNA padlock probing and in-situ sequencing.
academic.oup.com/nar/article/...
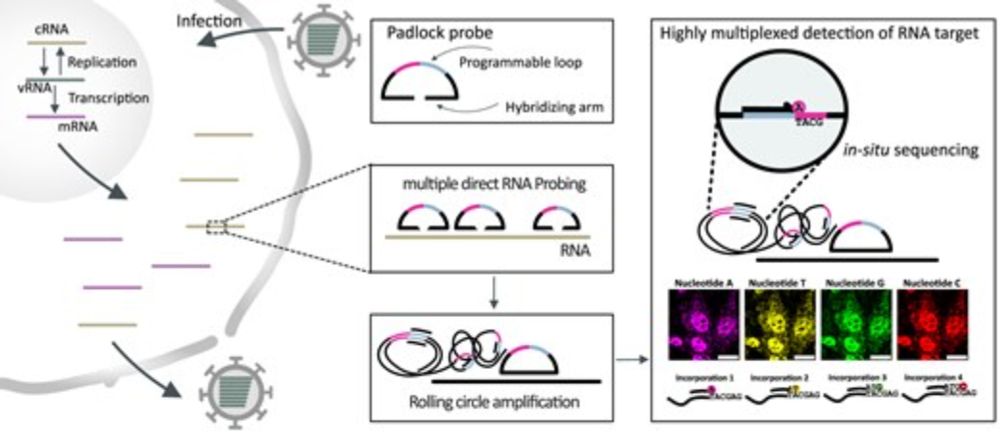
We established mudRapp-seq, a spatial transcriptomic technique that combines direct RNA padlock probing and in-situ sequencing.
academic.oup.com/nar/article/...
www.nature.com/articles/s41...
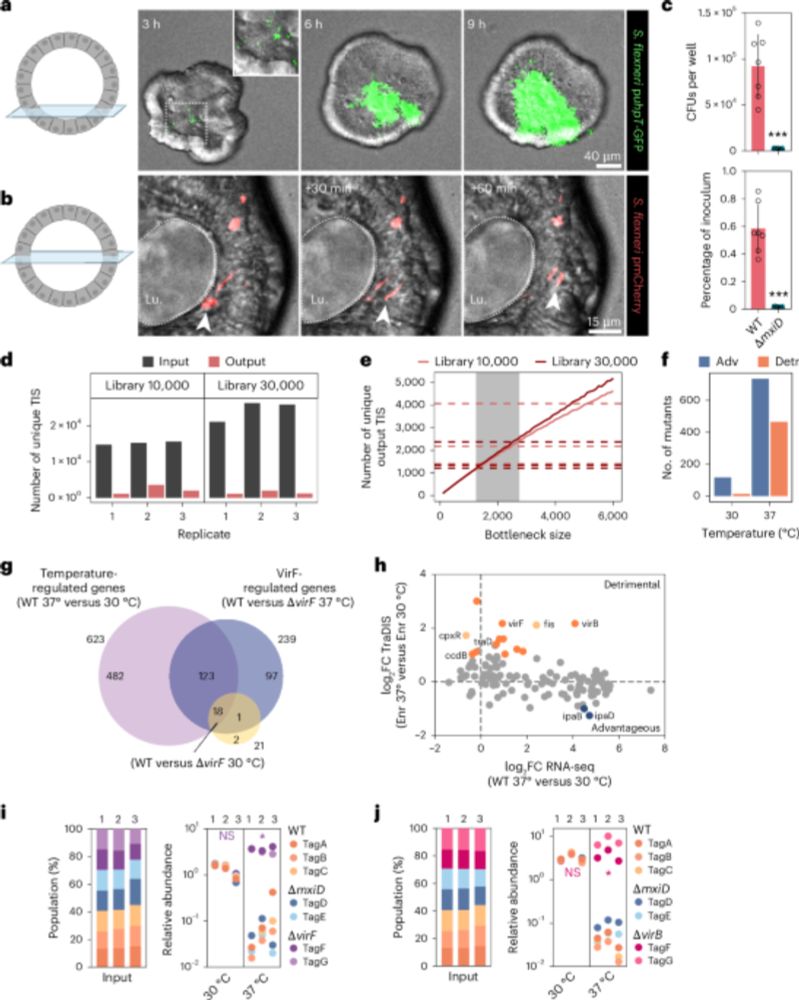
www.nature.com/articles/s41...
www.helmholtz-hzi.de/media-center...
www.helmholtz-hzi.de/media-center...

For a detailed protocol, see rdcu.be/eg0Xc, now available @natprot.bsky.social.
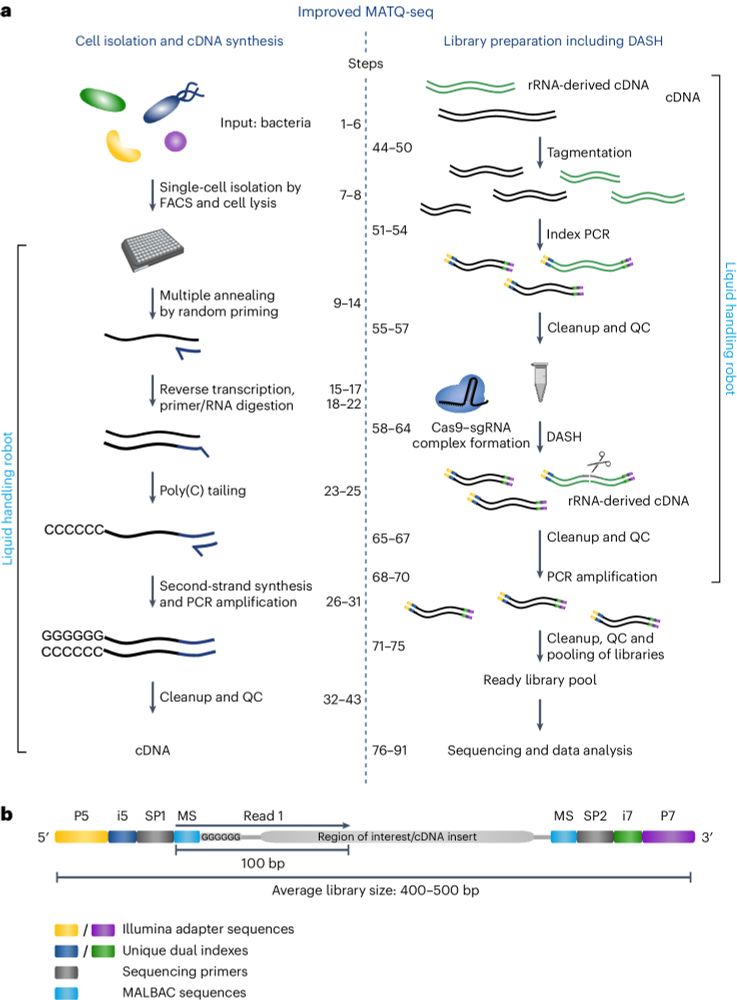
For a detailed protocol, see rdcu.be/eg0Xc, now available @natprot.bsky.social.


