https://www.helmholtz-hiri.de/en/research/associated-scientists-alumni/teams/team/host-pathogen-microbiota-interactions/

Congrats to @kooshapour.bsky.social & great collaboration w/ @jorg-vogel-lab.bsky.social!
doi.org/10.1093/nar/...

Congrats to @kooshapour.bsky.social & great collaboration w/ @jorg-vogel-lab.bsky.social!
doi.org/10.1093/nar/...



Our new study reveals the stepwise evolutionary journey of UhpU, a 3′UTR-derived sRNA in E. coli and relatives. It evolved from a metabolically-focused regulator to one with expanded targets and biogenesis manner.
www.pnas.org/doi/10.1073/...

Our new study reveals the stepwise evolutionary journey of UhpU, a 3′UTR-derived sRNA in E. coli and relatives. It evolved from a metabolically-focused regulator to one with expanded targets and biogenesis manner.
www.pnas.org/doi/10.1073/...
buff.ly/jJd9Eho

buff.ly/jJd9Eho
We developed new genetic tools & genome-wide libraries for species of the Bacteroidales order; constructed saturated barcoded transposon libraries in key representatives of three genera.
www.biorxiv.org/content/10.1...

We developed new genetic tools & genome-wide libraries for species of the Bacteroidales order; constructed saturated barcoded transposon libraries in key representatives of three genera.
www.biorxiv.org/content/10.1...

She focused on global RNA-binding proteins in Bacteroides—bacteria lacking Hfq, ProQ, CsrA, Khp—culminating in the discovery of a post-transcriptional network governed by RbpB (www.nature.com/articles/s41467-024-55383-8).

She focused on global RNA-binding proteins in Bacteroides—bacteria lacking Hfq, ProQ, CsrA, Khp—culminating in the discovery of a post-transcriptional network governed by RbpB (www.nature.com/articles/s41467-024-55383-8).

doi.org/10.1101/2025.09.08.672848

doi.org/10.1101/2025.09.08.672848

and @joaquinbernal.bsky.social : when macrophages are provided with specific nutrients, Salmonella can bypass its requirement of the T3SS to replicate, a process that occurs within the SCV
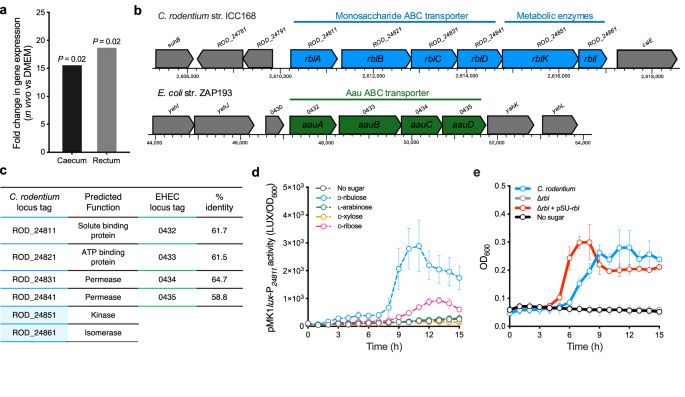
In this study lead by @lenaamend.bsky.social from the group, we explore carbon cross-feeding between a gut commensal and an enteropathogen. Can we counter infection by cutting sugar supply to pathogens?

In this study lead by @lenaamend.bsky.social from the group, we explore carbon cross-feeding between a gut commensal and an enteropathogen. Can we counter infection by cutting sugar supply to pathogens?
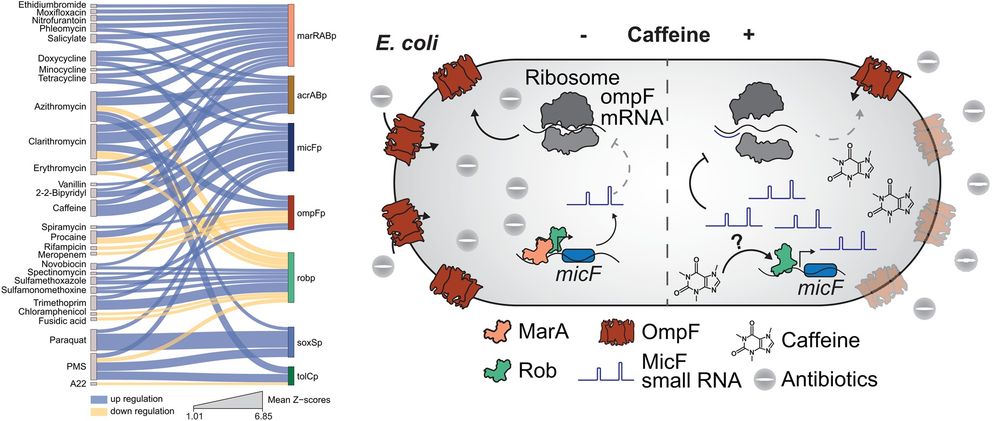
Our latest research is out @nature.com: We show that non-antibiotic drugs can disrupt colonization resistance, raising the risk of enteric infections.
rdcu.be/ewwrG
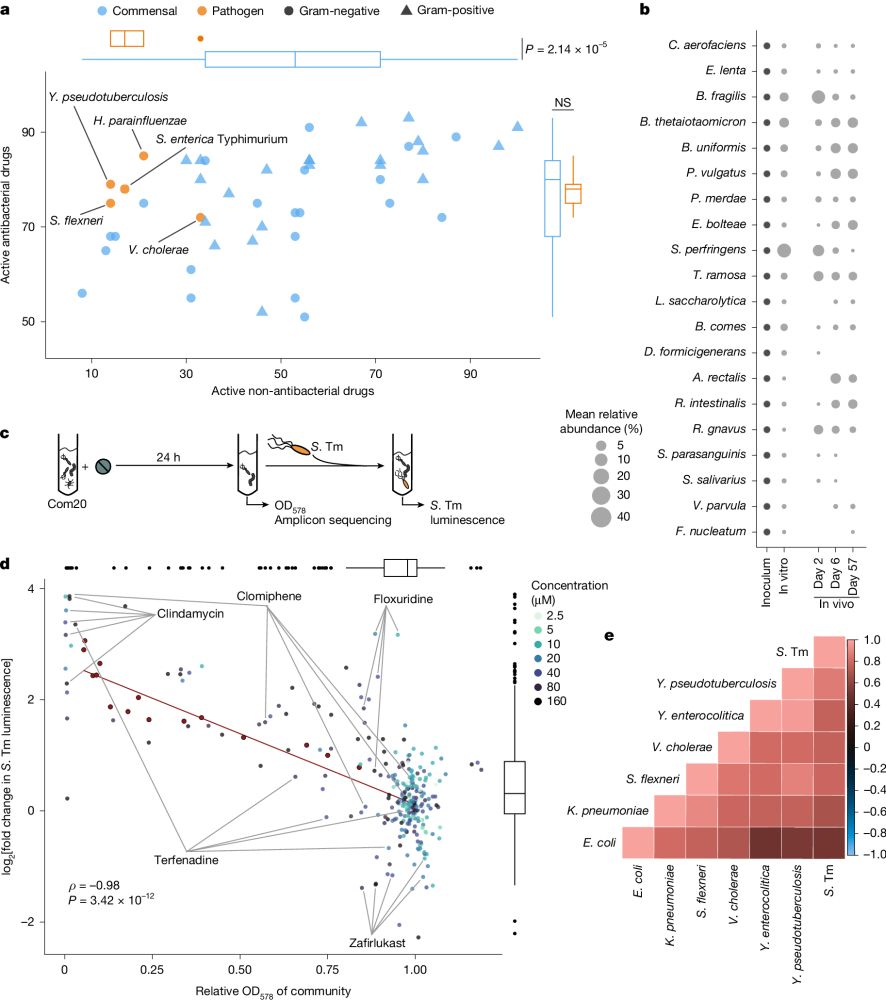
Our latest research is out @nature.com: We show that non-antibiotic drugs can disrupt colonization resistance, raising the risk of enteric infections.
rdcu.be/ewwrG
From 40 nominations, four top PIs were selected:
🥇 Gabriele Büchel
🥈 Alexander Westermann @westermannlab.bsky.social
🥉 Carmen Villmann & Matthias Gamer @gamerlab.bsky.social
Proud to have the best supervisors! @uni-wuerzburg.de

Ultra-sensitive RNA-seq and FISH show 𝘉𝘢𝘤𝘵𝘦𝘳𝘰𝘪𝘥𝘦𝘴 𝘵𝘩𝘦𝘵𝘢𝘪𝘰𝘵𝘢𝘰𝘮𝘪𝘤𝘳𝘰𝘯 cells adopt different shapes reflecting distinct gene expression profiles and metabolic specializations
A deliberate survival strategy for the ever-changing gut environment?
www.cell.com/cell-reports...
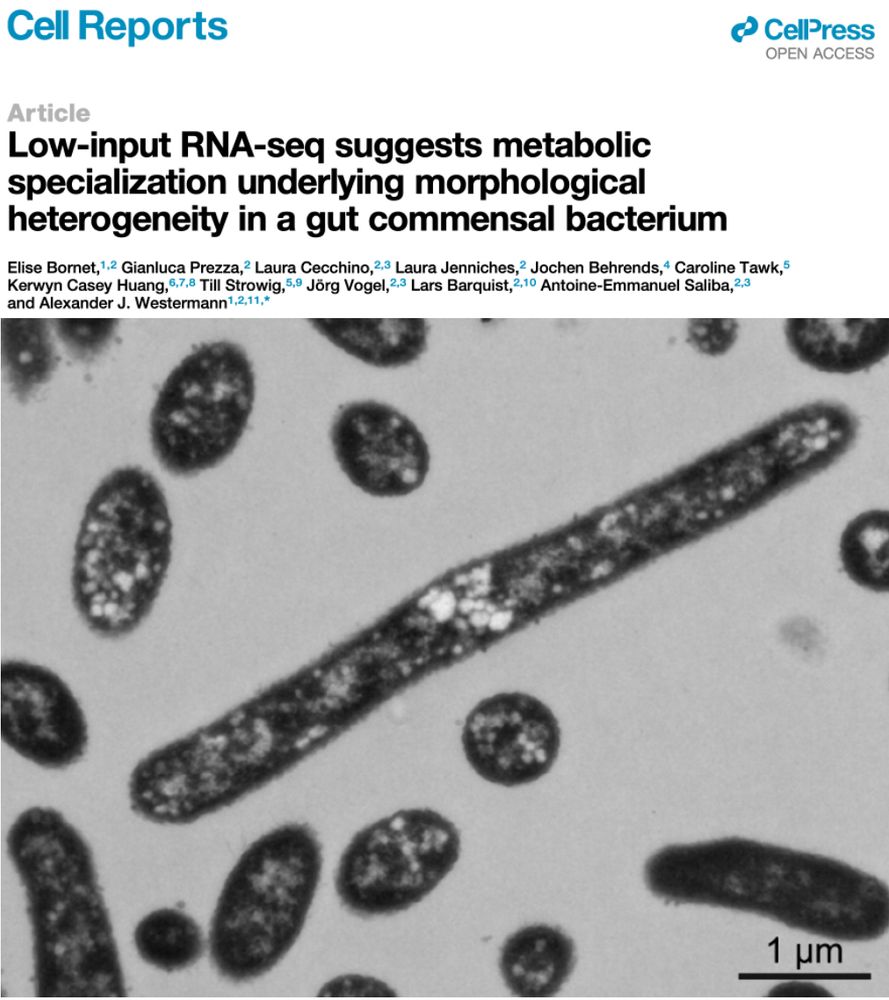
Ultra-sensitive RNA-seq and FISH show 𝘉𝘢𝘤𝘵𝘦𝘳𝘰𝘪𝘥𝘦𝘴 𝘵𝘩𝘦𝘵𝘢𝘪𝘰𝘵𝘢𝘰𝘮𝘪𝘤𝘳𝘰𝘯 cells adopt different shapes reflecting distinct gene expression profiles and metabolic specializations
A deliberate survival strategy for the ever-changing gut environment?
www.cell.com/cell-reports...

