Westermann Lab
@westermannlab.bsky.social
Microbiota RNA Lab at the University of Würzburg and the Helmholtz Institute for RNA-based Infection Research.
https://www.helmholtz-hiri.de/en/research/associated-scientists-alumni/teams/team/host-pathogen-microbiota-interactions/
https://www.helmholtz-hiri.de/en/research/associated-scientists-alumni/teams/team/host-pathogen-microbiota-interactions/
Reposted by Westermann Lab
De-DUFing the DUFs 🧩 @franznarberhaus.bsky.social lab uncovers how small DUF1127 proteins regulate #phosphate uptake by binding the sensor kinase PhoR. Their conserved role from Agrobacterium to E. coli highlights how even small DUFs can shape bacterial physiology 🦠
buff.ly/jJd9Eho
buff.ly/jJd9Eho

October 21, 2025 at 7:02 AM
De-DUFing the DUFs 🧩 @franznarberhaus.bsky.social lab uncovers how small DUF1127 proteins regulate #phosphate uptake by binding the sensor kinase PhoR. Their conserved role from Agrobacterium to E. coli highlights how even small DUFs can shape bacterial physiology 🦠
buff.ly/jJd9Eho
buff.ly/jJd9Eho
Reposted by Westermann Lab
Excited to share our preprint led by Carlos Voogdt et al
We developed new genetic tools & genome-wide libraries for species of the Bacteroidales order; constructed saturated barcoded transposon libraries in key representatives of three genera.
www.biorxiv.org/content/10.1...
We developed new genetic tools & genome-wide libraries for species of the Bacteroidales order; constructed saturated barcoded transposon libraries in key representatives of three genera.
www.biorxiv.org/content/10.1...

October 13, 2025 at 7:48 AM
Excited to share our preprint led by Carlos Voogdt et al
We developed new genetic tools & genome-wide libraries for species of the Bacteroidales order; constructed saturated barcoded transposon libraries in key representatives of three genera.
www.biorxiv.org/content/10.1...
We developed new genetic tools & genome-wide libraries for species of the Bacteroidales order; constructed saturated barcoded transposon libraries in key representatives of three genera.
www.biorxiv.org/content/10.1...
Reposted by Westermann Lab
How to search for unknown RNA-binding proteins? We did GradR in the cyanobacterium Synechocystis 6803. And now our paper „RAPDOR: Using Jensen-Shannon Distance for the computational analysis of complex proteomics dataset“ is finally out in Nature Communications, here: rdcu.be/eImcr .

RAPDOR: Using Jensen-Shannon Distance for the computational analysis of complex proteomics datasets
Nature Communications - RNA-binding proteins play key roles in post-transcriptional gene regulation. Here, Hemm et al. developed RAPDOR, a widely applicable tool based on Jensen-Shannon Distance...
rdcu.be
September 26, 2025 at 3:51 PM
How to search for unknown RNA-binding proteins? We did GradR in the cyanobacterium Synechocystis 6803. And now our paper „RAPDOR: Using Jensen-Shannon Distance for the computational analysis of complex proteomics dataset“ is finally out in Nature Communications, here: rdcu.be/eImcr .
Congrats to Ann-Sophie Rüttiger who successfully defended her PhD this week! 🎓
She focused on global RNA-binding proteins in Bacteroides—bacteria lacking Hfq, ProQ, CsrA, Khp—culminating in the discovery of a post-transcriptional network governed by RbpB (www.nature.com/articles/s41467-024-55383-8).
She focused on global RNA-binding proteins in Bacteroides—bacteria lacking Hfq, ProQ, CsrA, Khp—culminating in the discovery of a post-transcriptional network governed by RbpB (www.nature.com/articles/s41467-024-55383-8).

September 25, 2025 at 10:06 AM
Congrats to Ann-Sophie Rüttiger who successfully defended her PhD this week! 🎓
She focused on global RNA-binding proteins in Bacteroides—bacteria lacking Hfq, ProQ, CsrA, Khp—culminating in the discovery of a post-transcriptional network governed by RbpB (www.nature.com/articles/s41467-024-55383-8).
She focused on global RNA-binding proteins in Bacteroides—bacteria lacking Hfq, ProQ, CsrA, Khp—culminating in the discovery of a post-transcriptional network governed by RbpB (www.nature.com/articles/s41467-024-55383-8).
Reposted by Westermann Lab
Looking for a new approach to studying or eliminating phages? Check out our study introducing anti-phage ASOs (antisense oligos) out in @Nature today. nature.com/articles/s4158…

September 10, 2025 at 3:40 PM
Looking for a new approach to studying or eliminating phages? Check out our study introducing anti-phage ASOs (antisense oligos) out in @Nature today. nature.com/articles/s4158…
New preprint from the lab, in collaboration with Wenhan Zhu (U Vanderbilt): using dual RNA-seq during B. theta colonization of the host mucous layer, we identify IroR--an iron-response sRNA that tunes capsule expression and facilitates adaptation to iron limitation.
doi.org/10.1101/2025.09.08.672848
doi.org/10.1101/2025.09.08.672848

An RNA regulates iron homeostasis and host mucus colonization in Bacteroides thetaiotaomicron
Symbiotic bacteria in the human intestinal microbiota provide many pivotal functions to human health and occupy distinct biogeographic niches within the gut. Yet the molecular basis underlying niche-s...
doi.org
September 9, 2025 at 7:49 AM
New preprint from the lab, in collaboration with Wenhan Zhu (U Vanderbilt): using dual RNA-seq during B. theta colonization of the host mucous layer, we identify IroR--an iron-response sRNA that tunes capsule expression and facilitates adaptation to iron limitation.
doi.org/10.1101/2025.09.08.672848
doi.org/10.1101/2025.09.08.672848
Reposted by Westermann Lab
Starting the meeting microbes and RNA- Gerhart on stage - lifetime achievement Lecture - congrats Gerhart!

September 1, 2025 at 2:19 PM
Starting the meeting microbes and RNA- Gerhart on stage - lifetime achievement Lecture - congrats Gerhart!
Reposted by Westermann Lab
A Salmonella T3SS-2 mutant grows fine in spleen macrophages, contradicting tissue culture dogma (PMID: 23236281). This observation was largely ignored, but adding certain carbon sources rescues growth in cultured macrophages, hinting that T3SS-2 may be doing something entirely different in vivo.
New pre-print in the lab! Beautiful work by Francisco Garcia-Rodriguez in collab with @kamovalenz.bsky.social
and @joaquinbernal.bsky.social : when macrophages are provided with specific nutrients, Salmonella can bypass its requirement of the T3SS to replicate, a process that occurs within the SCV
and @joaquinbernal.bsky.social : when macrophages are provided with specific nutrients, Salmonella can bypass its requirement of the T3SS to replicate, a process that occurs within the SCV
Provision of Preferred Nutrients to Macrophages Enables Salmonella to Replicate Intracellularly Without Relying on Type III Secretion Systems https://www.biorxiv.org/content/10.1101/2025.05.15.653970v1
August 9, 2025 at 12:18 AM
A Salmonella T3SS-2 mutant grows fine in spleen macrophages, contradicting tissue culture dogma (PMID: 23236281). This observation was largely ignored, but adding certain carbon sources rescues growth in cultured macrophages, hinting that T3SS-2 may be doing something entirely different in vivo.
Latest preprint from the lab: doi.org/10.1101/2025...
In this study lead by @lenaamend.bsky.social from the group, we explore carbon cross-feeding between a gut commensal and an enteropathogen. Can we counter infection by cutting sugar supply to pathogens?
In this study lead by @lenaamend.bsky.social from the group, we explore carbon cross-feeding between a gut commensal and an enteropathogen. Can we counter infection by cutting sugar supply to pathogens?

Ablation of polysaccharide breakdown in Bacteroides thetaiotaomicron prevents cross-feeding and growth of Salmonella Typhimurium in the mouse gut
Pathogens invading the intestine compete for nutrients with the resident microbiota. However, there is evidence that commensal members of the gut also provide nutritional resources to enteropathogens ...
doi.org
August 17, 2025 at 6:13 AM
Latest preprint from the lab: doi.org/10.1101/2025...
In this study lead by @lenaamend.bsky.social from the group, we explore carbon cross-feeding between a gut commensal and an enteropathogen. Can we counter infection by cutting sugar supply to pathogens?
In this study lead by @lenaamend.bsky.social from the group, we explore carbon cross-feeding between a gut commensal and an enteropathogen. Can we counter infection by cutting sugar supply to pathogens?
Reposted by Westermann Lab
Absolutely delighted to share the labs latest work. We identify and characterise two pathways for D-ribulose utilisation in pathogenic EHEC and Citrobacter.
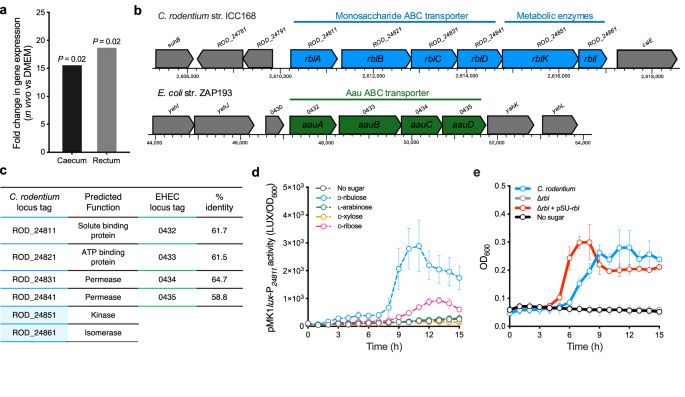
Convergent evolution of distinct D-ribulose utilisation pathways in attaching and effacing pathogens - Nature Communications
Cottam et al. identify distinct pathways for D-ribulose utilisation in pathogenic Escherichia coli and Citrobacter rodentium, providing mechanistic details and suggesting convergent evolution towards ...
www.nature.com
July 30, 2025 at 9:24 AM
Absolutely delighted to share the labs latest work. We identify and characterise two pathways for D-ribulose utilisation in pathogenic EHEC and Citrobacter.
Reposted by Westermann Lab
Finally online!
Our latest research is out @nature.com: We show that non-antibiotic drugs can disrupt colonization resistance, raising the risk of enteric infections.
rdcu.be/ewwrG
Our latest research is out @nature.com: We show that non-antibiotic drugs can disrupt colonization resistance, raising the risk of enteric infections.
rdcu.be/ewwrG
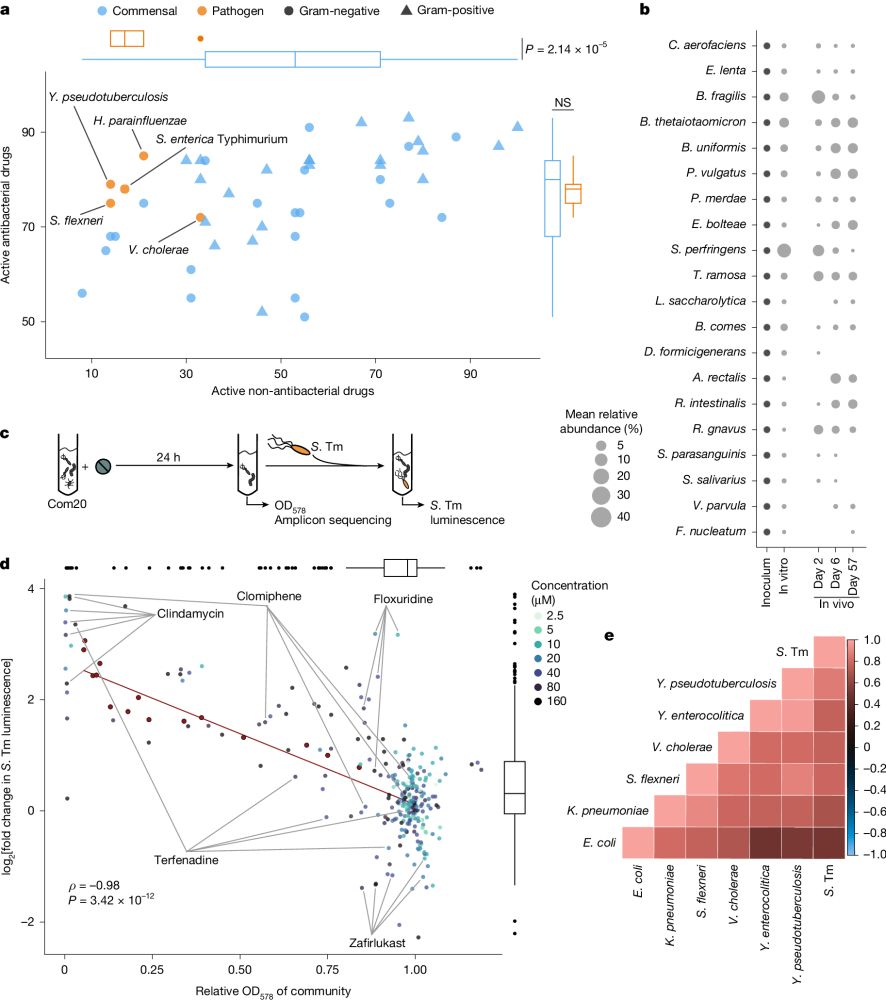
Non-antibiotics disrupt colonization resistance against enteropathogens
Nature - Non-antibiotic drugs from a wide range of therapeutic classes can alter the ability of gut commensals to resist invasion by enteropathogens, a previously underappreciated side effect of...
rdcu.be
July 16, 2025 at 9:20 PM
Finally online!
Our latest research is out @nature.com: We show that non-antibiotic drugs can disrupt colonization resistance, raising the risk of enteric infections.
rdcu.be/ewwrG
Our latest research is out @nature.com: We show that non-antibiotic drugs can disrupt colonization resistance, raising the risk of enteric infections.
rdcu.be/ewwrG
Reposted by Westermann Lab
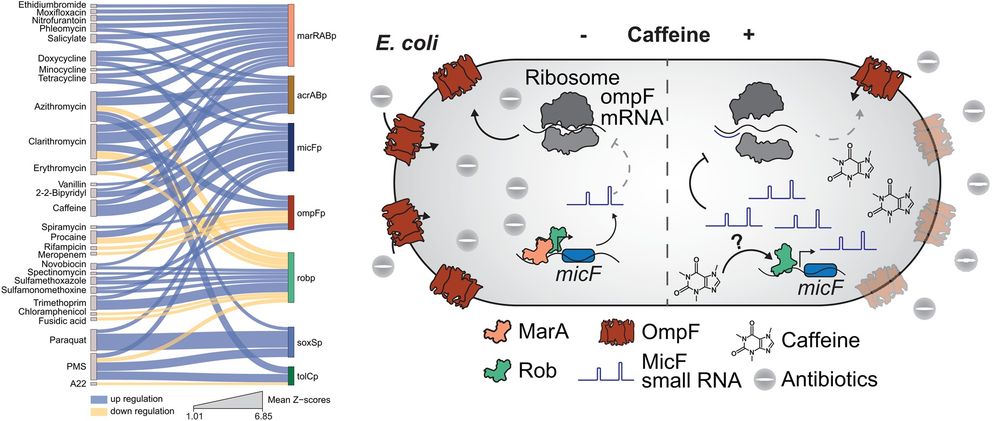
How are bacterial transport machineries regulated by external cues? @brochadolab.bsky.social &co identify environmental chemicals that regulate #transporter transcription, identifying the TF Rob as a key modulator of #EffluxPump expression in #Ecoli #AMR @plosbiology.org 🧪 plos.io/46pUM9Q
July 23, 2025 at 9:20 AM
How are bacterial transport machineries regulated by external cues? @brochadolab.bsky.social &co identify environmental chemicals that regulate #transporter transcription, identifying the TF Rob as a key modulator of #EffluxPump expression in #Ecoli #AMR @plosbiology.org 🧪 plos.io/46pUM9Q
Reposted by Westermann Lab
New preprint! We characterise the small, regulatory RNA Arp which controls DNA uptake and twitching motilty in A. baumannii. Led by our Dr Fergal Hamrock @hamrockfergal.bsky.social and in collaboration with @westermannlab.bsky.social and Mike Gebhardt's lab!
DNA uptake and twitching motility are controlled by the small RNA Arp through repression of pilin translation in Acinetobacter baumannii https://www.biorxiv.org/content/10.1101/2025.07.19.665661v1
July 21, 2025 at 2:56 PM
New preprint! We characterise the small, regulatory RNA Arp which controls DNA uptake and twitching motilty in A. baumannii. Led by our Dr Fergal Hamrock @hamrockfergal.bsky.social and in collaboration with @westermannlab.bsky.social and Mike Gebhardt's lab!
Congrats to Gianluca Prezza (@gprezza.bsky.social) for reveiving the 2025 PhD Award from the Friends of the Helmholtz Centre for Infection Research (HZI)!👏 The award honours excellent PhD theses from the field of life sciences at HZI partner universities. His thesis focused Bacteroides' RNA biology.
Beim gestrigen Festakt wurden zudem die HZI-Promotionspreise an Wenchao Li @ciim-hannover.bsky.social und @gprezza.bsky.social aus der Gruppe von @westermannlab.bsky.social @helmholtz-hiri.bsky.social verliehen. Wir gratulieren den beiden zu ihren ausgezeichneten Doktorarbeiten!
July 11, 2025 at 9:35 AM
Congrats to Gianluca Prezza (@gprezza.bsky.social) for reveiving the 2025 PhD Award from the Friends of the Helmholtz Centre for Infection Research (HZI)!👏 The award honours excellent PhD theses from the field of life sciences at HZI partner universities. His thesis focused Bacteroides' RNA biology.
Very honored that our recent study on Bacteroides morphotypes was spotlighted 🔦 in Trends in Microbiology by Eric Martens & Qinnan Yang: www.cell.com/trends/micro....
Low biomass bacterial transcriptomics takes shape
Bornet et al. apply a low-input bacterial RNA-seq pipeline to transcriptionally profile
small, medium, and large cell populations of the human gut symbiont Bacteroides thetaiotaomicron
separated by fl...
www.cell.com
July 8, 2025 at 10:59 AM
Very honored that our recent study on Bacteroides morphotypes was spotlighted 🔦 in Trends in Microbiology by Eric Martens & Qinnan Yang: www.cell.com/trends/micro....
Thank you to all students and to the GSLS for this award! It means a lot to me! (-AW)
We proudly present the first-ever #GSLS Supervisor of the Year Award! 🏆
From 40 nominations, four top PIs were selected:
🥇 Gabriele Büchel
🥈 Alexander Westermann @westermannlab.bsky.social
🥉 Carmen Villmann & Matthias Gamer @gamerlab.bsky.social
Proud to have the best supervisors! @uni-wuerzburg.de
From 40 nominations, four top PIs were selected:
🥇 Gabriele Büchel
🥈 Alexander Westermann @westermannlab.bsky.social
🥉 Carmen Villmann & Matthias Gamer @gamerlab.bsky.social
Proud to have the best supervisors! @uni-wuerzburg.de

July 7, 2025 at 7:46 AM
Thank you to all students and to the GSLS for this award! It means a lot to me! (-AW)
Reposted by Westermann Lab
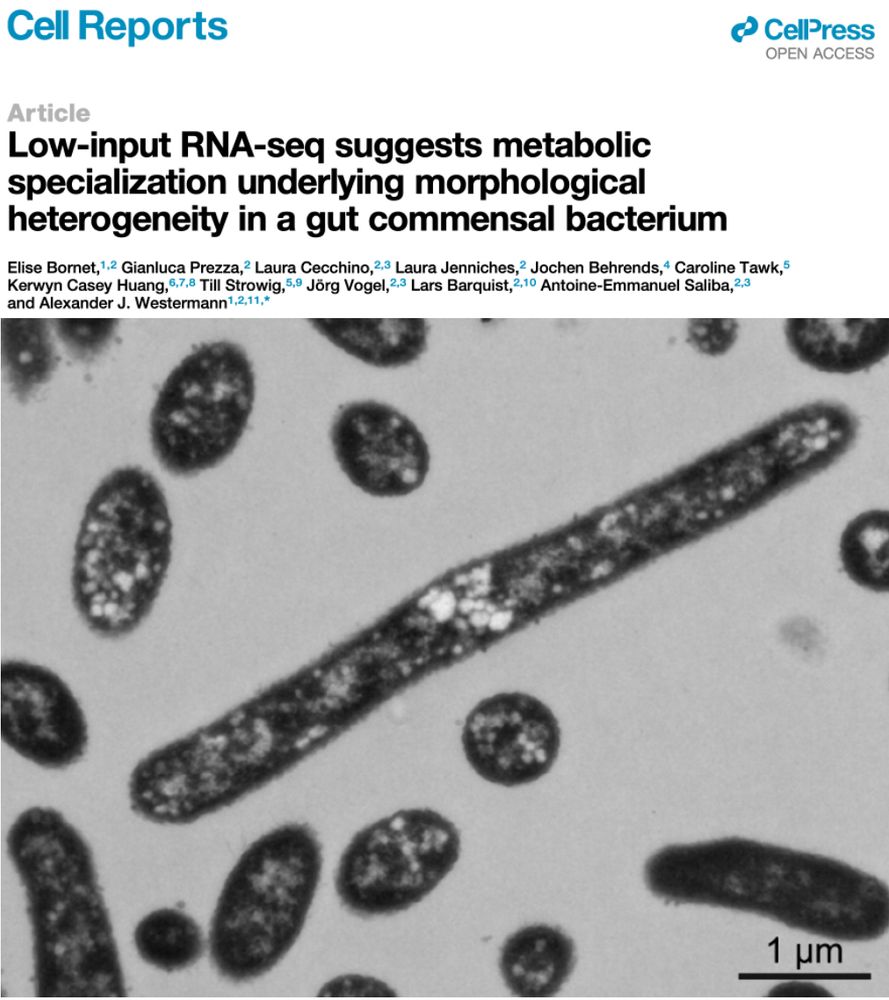
𝔟α𝐜𝐓𝕖Ř𝒾ⒶĹ SHⓐ𝓹Ẹ 丂ħ𝓲ƒ𝓉ⒺⓇร 🦠
Ultra-sensitive RNA-seq and FISH show 𝘉𝘢𝘤𝘵𝘦𝘳𝘰𝘪𝘥𝘦𝘴 𝘵𝘩𝘦𝘵𝘢𝘪𝘰𝘵𝘢𝘰𝘮𝘪𝘤𝘳𝘰𝘯 cells adopt different shapes reflecting distinct gene expression profiles and metabolic specializations
A deliberate survival strategy for the ever-changing gut environment?
www.cell.com/cell-reports...
Ultra-sensitive RNA-seq and FISH show 𝘉𝘢𝘤𝘵𝘦𝘳𝘰𝘪𝘥𝘦𝘴 𝘵𝘩𝘦𝘵𝘢𝘪𝘰𝘵𝘢𝘰𝘮𝘪𝘤𝘳𝘰𝘯 cells adopt different shapes reflecting distinct gene expression profiles and metabolic specializations
A deliberate survival strategy for the ever-changing gut environment?
www.cell.com/cell-reports...
June 18, 2025 at 1:02 AM
𝔟α𝐜𝐓𝕖Ř𝒾ⒶĹ SHⓐ𝓹Ẹ 丂ħ𝓲ƒ𝓉ⒺⓇร 🦠
Ultra-sensitive RNA-seq and FISH show 𝘉𝘢𝘤𝘵𝘦𝘳𝘰𝘪𝘥𝘦𝘴 𝘵𝘩𝘦𝘵𝘢𝘪𝘰𝘵𝘢𝘰𝘮𝘪𝘤𝘳𝘰𝘯 cells adopt different shapes reflecting distinct gene expression profiles and metabolic specializations
A deliberate survival strategy for the ever-changing gut environment?
www.cell.com/cell-reports...
Ultra-sensitive RNA-seq and FISH show 𝘉𝘢𝘤𝘵𝘦𝘳𝘰𝘪𝘥𝘦𝘴 𝘵𝘩𝘦𝘵𝘢𝘪𝘰𝘵𝘢𝘰𝘮𝘪𝘤𝘳𝘰𝘯 cells adopt different shapes reflecting distinct gene expression profiles and metabolic specializations
A deliberate survival strategy for the ever-changing gut environment?
www.cell.com/cell-reports...
Our latest: www.cell.com/cell-reports/fulltext/S2211-1247(25)00615-1. Links Bacteroides cell size w/ gene expression. Protocol works even for single cells. 🦠
Shoutout to @elisebor.bsky.social, @emmanuel-saliba.bsky.social, @lbarquist.bsky.social, @jorg-vogel-lab.bsky.social, Till Strowig & KC Huang.
Shoutout to @elisebor.bsky.social, @emmanuel-saliba.bsky.social, @lbarquist.bsky.social, @jorg-vogel-lab.bsky.social, Till Strowig & KC Huang.
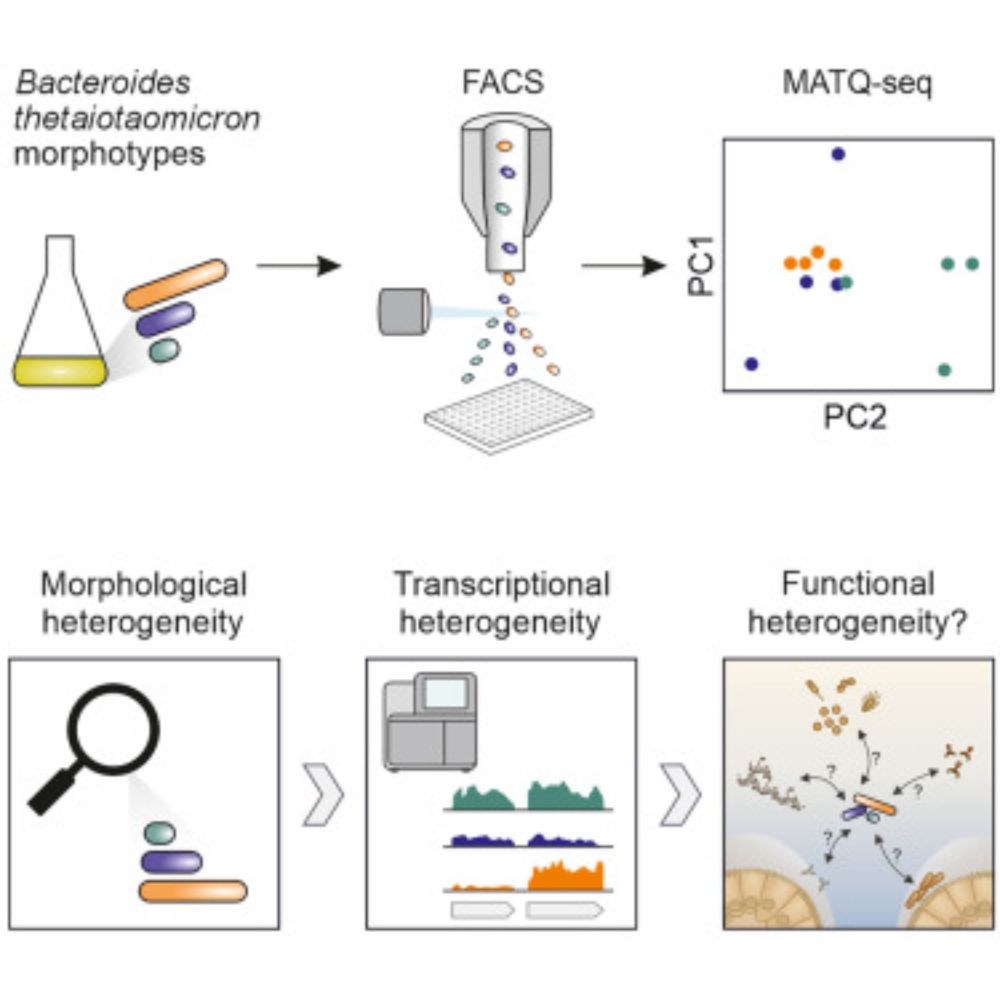
Low-input RNA-seq suggests metabolic specialization underlying morphological heterogeneity in a gut commensal bacterium
Bornet et al. investigate the morphological heterogeneity of Bacteroides thetaiotaomicron
cells observed both in laboratory culture and inside the mammalian GI tract. The authors
show that cell size i...
www.cell.com
June 17, 2025 at 7:11 PM
Our latest: www.cell.com/cell-reports/fulltext/S2211-1247(25)00615-1. Links Bacteroides cell size w/ gene expression. Protocol works even for single cells. 🦠
Shoutout to @elisebor.bsky.social, @emmanuel-saliba.bsky.social, @lbarquist.bsky.social, @jorg-vogel-lab.bsky.social, Till Strowig & KC Huang.
Shoutout to @elisebor.bsky.social, @emmanuel-saliba.bsky.social, @lbarquist.bsky.social, @jorg-vogel-lab.bsky.social, Till Strowig & KC Huang.
Reposted by Westermann Lab
Very excited about our latest combining advanced cell culture, transposon-insertion sequencing, and statistical modeling to map Shigella virulence determinants in organoid models. Great collaboration w/ @mldm.bsky.social @sellinlab.bsky.social in @natgenet.nature.com
www.nature.com/articles/s41...
www.nature.com/articles/s41...
A scalable gut epithelial organoid model reveals the genome-wide colonization landscape of a human-adapted pathogen - Nature Genetics
A genome-wide screen using human gut epithelial organoids combined with transposon-directed insertion sequencing identifies over 100 Shigella flexneri genes required for epithelial colonization.
www.nature.com
June 12, 2025 at 1:19 PM
Very excited about our latest combining advanced cell culture, transposon-insertion sequencing, and statistical modeling to map Shigella virulence determinants in organoid models. Great collaboration w/ @mldm.bsky.social @sellinlab.bsky.social in @natgenet.nature.com
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Reposted by Westermann Lab
Bacterial genomes still contain many hidden genes.
This article reports a high-resolution map of translated proteins, incl. small ones, in the pathogen Campylobacter jejuni
@uniwuerzburg.bsky.social @campy-bara.bsky.social @sharmalab.bsky.social
This article reports a high-resolution map of translated proteins, incl. small ones, in the pathogen Campylobacter jejuni
@uniwuerzburg.bsky.social @campy-bara.bsky.social @sharmalab.bsky.social
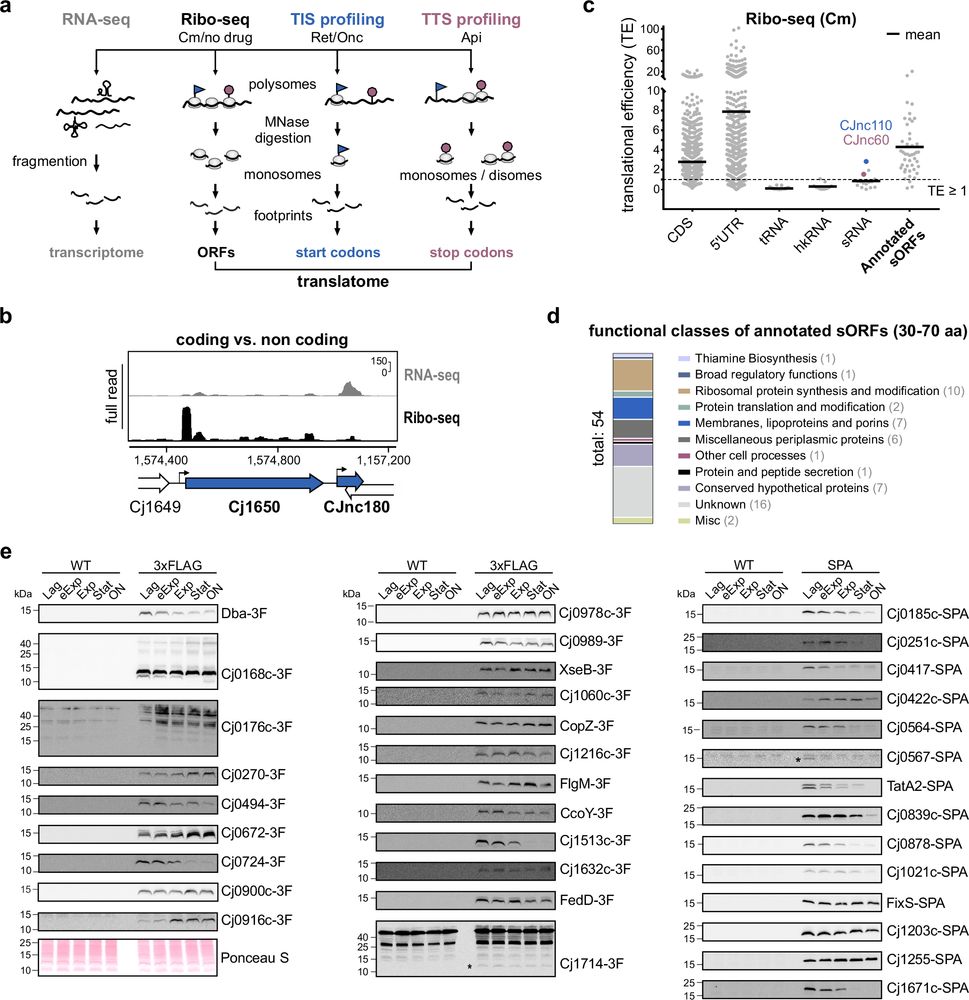
Complementary Ribo-seq approaches map the translatome and provide a small protein census in the foodborne pathogen Campylobacter jejuni - Nature Communications
Bacterial genomes still contain many hidden and uncharacterized genes. Using complementary Ribo-seq approaches, Froschauer, Svensson et al. here annotate start and stop codons of Campylobacter jejuni, reporting a high-resolution translatome and identifying several novel small ORFs in this foodborne pathogen.
bit.ly
May 28, 2025 at 10:45 AM
Bacterial genomes still contain many hidden genes.
This article reports a high-resolution map of translated proteins, incl. small ones, in the pathogen Campylobacter jejuni
@uniwuerzburg.bsky.social @campy-bara.bsky.social @sharmalab.bsky.social
This article reports a high-resolution map of translated proteins, incl. small ones, in the pathogen Campylobacter jejuni
@uniwuerzburg.bsky.social @campy-bara.bsky.social @sharmalab.bsky.social
Reposted by Westermann Lab
Erfolg auf ganzer Linie: zwei Exzellenzcluster für Würzburg! 🥂🚀
Im Rahmen der #ExStra wird nicht nur der Quantenphysik-Cluster ctd.qmat weiterhin gefördert. Mit NUCLEATE, einem Forschungsprojekt zu Nukleinsäuren, kommt noch ein zweiter Cluster hinzu!
➡️ go.uniwue.de/xstra25
Im Rahmen der #ExStra wird nicht nur der Quantenphysik-Cluster ctd.qmat weiterhin gefördert. Mit NUCLEATE, einem Forschungsprojekt zu Nukleinsäuren, kommt noch ein zweiter Cluster hinzu!
➡️ go.uniwue.de/xstra25

May 22, 2025 at 4:49 PM
Erfolg auf ganzer Linie: zwei Exzellenzcluster für Würzburg! 🥂🚀
Im Rahmen der #ExStra wird nicht nur der Quantenphysik-Cluster ctd.qmat weiterhin gefördert. Mit NUCLEATE, einem Forschungsprojekt zu Nukleinsäuren, kommt noch ein zweiter Cluster hinzu!
➡️ go.uniwue.de/xstra25
Im Rahmen der #ExStra wird nicht nur der Quantenphysik-Cluster ctd.qmat weiterhin gefördert. Mit NUCLEATE, einem Forschungsprojekt zu Nukleinsäuren, kommt noch ein zweiter Cluster hinzu!
➡️ go.uniwue.de/xstra25
Reposted by Westermann Lab
And this only the beginning! I’m glad we did that collaboration with the Maric lab. ASOs for the people!
First post, first paper 🤭
Proud to share with all of you our work on mRNA blocking using our own high-throughput antisense approach! doi.org/10.1002/advs...
Many thanks to @hmariclab.bsky.social, @jorg-vogel-lab.bsky.social and especially Linda for the amazing collaboration 😸
Proud to share with all of you our work on mRNA blocking using our own high-throughput antisense approach! doi.org/10.1002/advs...
Many thanks to @hmariclab.bsky.social, @jorg-vogel-lab.bsky.social and especially Linda for the amazing collaboration 😸
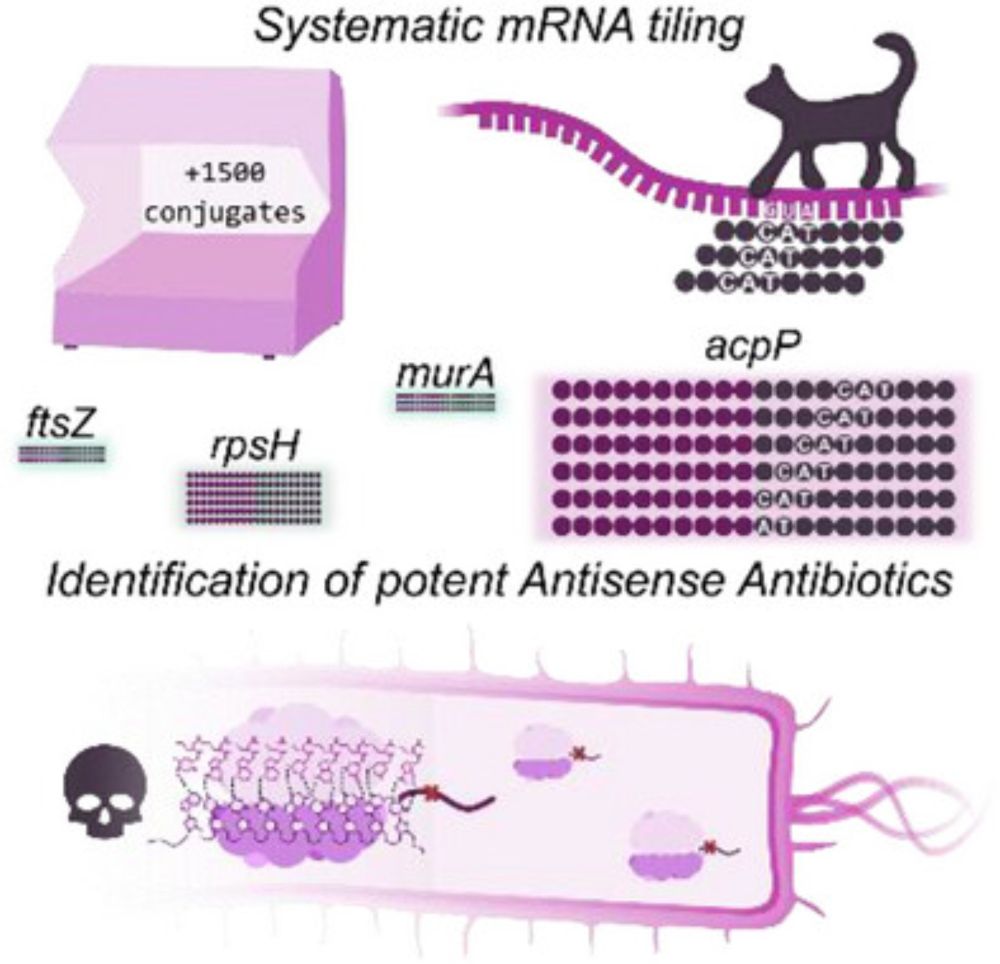
High‐Throughput Tiling of Essential mRNAs Increases Potency of Antisense Antibiotics
The systematic tiling of essential genes’ mRNA here presented, proposes a valuable tool for the identification of novel PNA sequences with antibiotic potential. The high-throughput synthetic set up o...
doi.org
May 5, 2025 at 2:00 PM
And this only the beginning! I’m glad we did that collaboration with the Maric lab. ASOs for the people!
Reposted by Westermann Lab
Here we share thoughts on next-gen cultivation: rdcu.be/ejPSb. Hope it helps 😄 Was fun to write with great colleagues @lisamaierlab.bsky.social @jselkrig.bsky.social @baerboletta.bsky.social @typaslab.bsky.social @mmmicrobiomelab.bsky.social @leibniz-dsmz.bsky.social @westermannlab.bsky.social
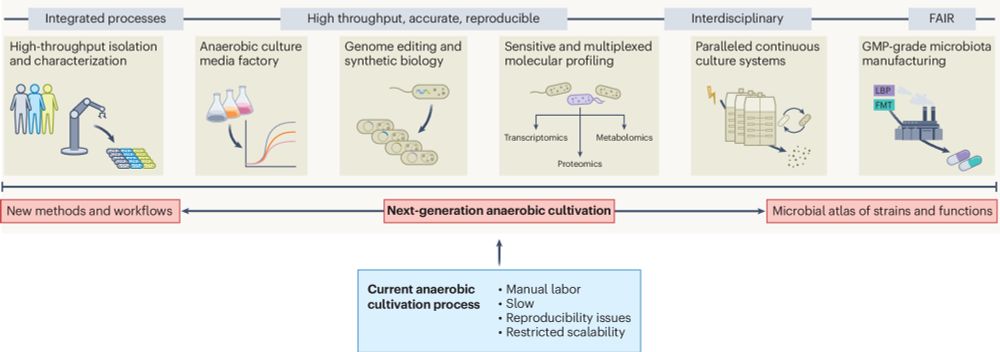
Enabling next-generation anaerobic cultivation through biotechnology to advance functional microbiome research
Nature Biotechnology - Overcoming key hurdles for the development of next-generation anaerobic cultivation methods could have major impacts on microbial community research and applications.
rdcu.be
April 29, 2025 at 11:58 AM
Here we share thoughts on next-gen cultivation: rdcu.be/ejPSb. Hope it helps 😄 Was fun to write with great colleagues @lisamaierlab.bsky.social @jselkrig.bsky.social @baerboletta.bsky.social @typaslab.bsky.social @mmmicrobiomelab.bsky.social @leibniz-dsmz.bsky.social @westermannlab.bsky.social
Reposted by Westermann Lab
I'm happy to share the first publication from my lab @hoerlab.bsky.social, now out in @narjournal.bsky.social! I take a look at the biology of RNA phages, focusing on how meta-omics methods are discovering new RNA phage families and bacterial defense strategies.
#PhageSky
doi.org/10.1093/nar/...
#PhageSky
doi.org/10.1093/nar/...
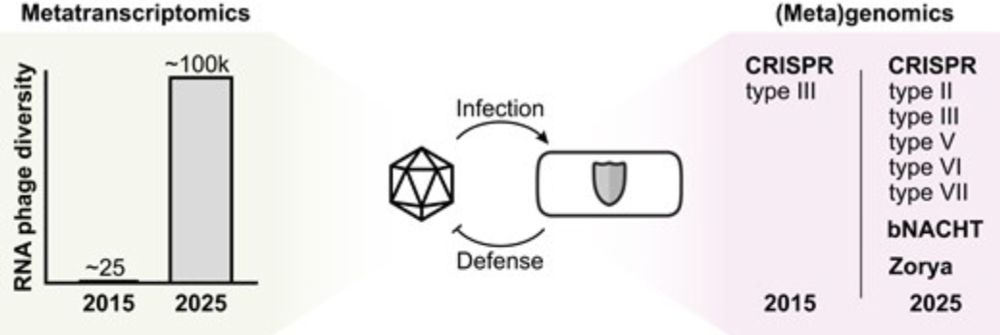
Advancing RNA phage biology through meta-omics
Abstract. Bacteriophages with RNA genomes are among the simplest biological entities on Earth. Since their discovery in the 1960s, they have been used as i
doi.org
April 23, 2025 at 8:57 AM
I'm happy to share the first publication from my lab @hoerlab.bsky.social, now out in @narjournal.bsky.social! I take a look at the biology of RNA phages, focusing on how meta-omics methods are discovering new RNA phage families and bacterial defense strategies.
#PhageSky
doi.org/10.1093/nar/...
#PhageSky
doi.org/10.1093/nar/...

