https://orcid.org/0000-0002-1054-2124
https://sites.temple.edu/voelzlab
Learn more here: buff.ly/gInH6VV
#SBGrid #SBGridSoftware #StructuralBiology

Learn more here: buff.ly/gInH6VV
#SBGrid #SBGridSoftware #StructuralBiology
Experiment-guided AlphaFold3 resolves accurate protein ensembles.
doi.org/10.1101/2025...
AlphaFold3 is incredible, but has crucial limitations: it typically collapses to a single conformation, ignoring the inherent dynamics of proteins. And it can be wrong. Here's a solution. 🧵👇

Experiment-guided AlphaFold3 resolves accurate protein ensembles.
doi.org/10.1101/2025...
AlphaFold3 is incredible, but has crucial limitations: it typically collapses to a single conformation, ignoring the inherent dynamics of proteins. And it can be wrong. Here's a solution. 🧵👇
Check out this new interview with Pat in Nature Reviews Drug Discovery:
www.nature.com/articles/d41...

Check out this new interview with Pat in Nature Reviews Drug Discovery:
www.nature.com/articles/d41...

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
pubs.acs.org/doi/10.1021/...

pubs.acs.org/doi/10.1021/...
from www.linkedin.com/in/ullah-sam...
www.youtube.com/watch?v=A5ph...
PyPI pypi.org/project/ipsae/
His github fork github.com/ullahsamee/I...
My github github.com/DunbrackLab/...
Paper www.biorxiv.org/content/10.1...
For designed protein binders www.biorxiv.org/content/10.1...

from www.linkedin.com/in/ullah-sam...
www.youtube.com/watch?v=A5ph...
PyPI pypi.org/project/ipsae/
His github fork github.com/ullahsamee/I...
My github github.com/DunbrackLab/...
Paper www.biorxiv.org/content/10.1...
For designed protein binders www.biorxiv.org/content/10.1...
Named after Quetzalcoatl, the Aztec god of creation, Quetzal is a simple yet scalable model for building 3D molecules atom by atom.
📜 arxiv.org/abs/2505.13791
[1/4]
Named after Quetzalcoatl, the Aztec god of creation, Quetzal is a simple yet scalable model for building 3D molecules atom by atom.
📜 arxiv.org/abs/2505.13791
[1/4]
www.nature.com/articles/s41...
www.nature.com/articles/s41...
We used molecular dynamics simulations and solid-state NMR (@fmp-berlin.de) to look at calcium binding to the pore domain of the calcium-gated K+ channel MthK. @wojciechkopec.bsky.social @compbiophys.bsky.social
We used molecular dynamics simulations and solid-state NMR (@fmp-berlin.de) to look at calcium binding to the pore domain of the calcium-gated K+ channel MthK. @wojciechkopec.bsky.social @compbiophys.bsky.social
🔗 doi.org/10.1021/acs....

🔗 doi.org/10.1021/acs....
The winners are being acknowledged for their outstanding achievements in chemistry across various fields in the discipline. cen.acs.org/people/award...

The winners are being acknowledged for their outstanding achievements in chemistry across various fields in the discipline. cen.acs.org/people/award...
bit.ly/4lnVYOs

bit.ly/4lnVYOs
Link: www.nature.com/articles/s41...

Link: www.nature.com/articles/s41...

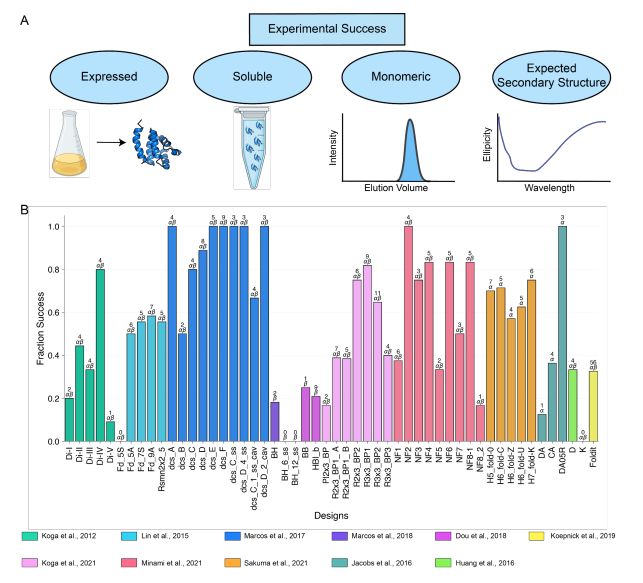
- deep learning structural metrics only weakly predict success
- The score distribution is different for different types of structures
@grocklin.bsky.social



- deep learning structural metrics only weakly predict success
- The score distribution is different for different types of structures
@grocklin.bsky.social
www.nature.com/articles/s41...
This technology, 11-years in the making, has transformed how we do research in my lab, and was led by an amazing postdoc, Matt Styles, who is currently entering the academic job market - so look out for him! 1/n
www.nature.com/articles/s41...
This technology, 11-years in the making, has transformed how we do research in my lab, and was led by an amazing postdoc, Matt Styles, who is currently entering the academic job market - so look out for him! 1/n
Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵

Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵


www.cecam.org/workshop-det...

