
Kevin K. Yang 楊凱筌
@kevinkaichuang.bsky.social
Principal Researcher in BioML at Microsoft Research. He/him/他. 🇹🇼 yangkky.github.io
Pinned
Three BioML starter packs now!
Pack 1: go.bsky.app/2VWBcCd
Pack 2: go.bsky.app/Bw84Hmc
Pack 3: go.bsky.app/NAKYUok
DM if you want to be included (or nominate people who should be!)
Pack 1: go.bsky.app/2VWBcCd
Pack 2: go.bsky.app/Bw84Hmc
Pack 3: go.bsky.app/NAKYUok
DM if you want to be included (or nominate people who should be!)
Design stable, folded proteins using only the 10 "ancient" amino acids.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...


October 31, 2025 at 7:40 PM
Design stable, folded proteins using only the 10 "ancient" amino acids.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
You should apply to do an internship with us!
- Make sure you attach a research statement (we don't consider anybody with no statement for interviews)
- The application site will probably go down for a day or two on Nov 17, and the link may change
jobs.careers.microsoft.com/global/en/jo...
- Make sure you attach a research statement (we don't consider anybody with no statement for interviews)
- The application site will probably go down for a day or two on Nov 17, and the link may change
jobs.careers.microsoft.com/global/en/jo...
October 31, 2025 at 2:11 PM
You should apply to do an internship with us!
- Make sure you attach a research statement (we don't consider anybody with no statement for interviews)
- The application site will probably go down for a day or two on Nov 17, and the link may change
jobs.careers.microsoft.com/global/en/jo...
- Make sure you attach a research statement (we don't consider anybody with no statement for interviews)
- The application site will probably go down for a day or two on Nov 17, and the link may change
jobs.careers.microsoft.com/global/en/jo...
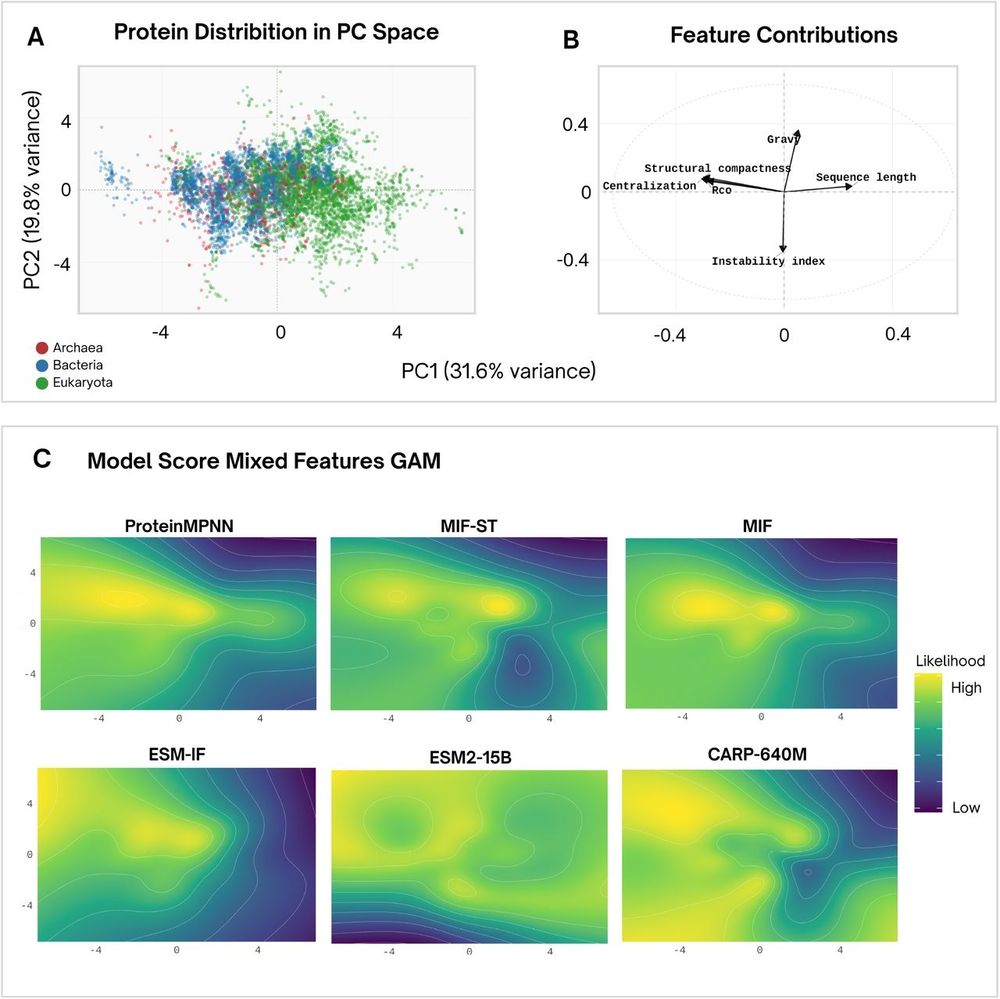
Structure - > sequence models have different phylogenetic (and biochemical) preferences than protein language models.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...



October 29, 2025 at 9:59 PM
Structure - > sequence models have different phylogenetic (and biochemical) preferences than protein language models.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Kevin K. Yang 楊凱筌
Are you a PhD student interested in ML and biology or health? Come do an internship with me, @avapamini.bsky.social, Alex Lu, @lcrawford.bsky.social, or Kristen Severson at MSRNE!
Applications are due Dec 1: make sure you include a research statement!
jobs.careers.microsoft.com/global/en/jo...
Applications are due Dec 1: make sure you include a research statement!
jobs.careers.microsoft.com/global/en/jo...
Search Jobs | Microsoft Careers
jobs.careers.microsoft.com
October 21, 2025 at 7:32 PM
Are you a PhD student interested in ML and biology or health? Come do an internship with me, @avapamini.bsky.social, Alex Lu, @lcrawford.bsky.social, or Kristen Severson at MSRNE!
Applications are due Dec 1: make sure you include a research statement!
jobs.careers.microsoft.com/global/en/jo...
Applications are due Dec 1: make sure you include a research statement!
jobs.careers.microsoft.com/global/en/jo...
Reposted by Kevin K. Yang 楊凱筌
Come do an internship with us! Please apply
Are you a PhD student interested in ML and biology or health? Come do an internship with me, @avapamini.bsky.social, Alex Lu, @lcrawford.bsky.social, or Kristen Severson at MSRNE!
Applications are due Dec 1: make sure you include a research statement!
jobs.careers.microsoft.com/global/en/jo...
Applications are due Dec 1: make sure you include a research statement!
jobs.careers.microsoft.com/global/en/jo...
Search Jobs | Microsoft Careers
jobs.careers.microsoft.com
October 22, 2025 at 12:41 PM
Come do an internship with us! Please apply
Are you a PhD student interested in ML and biology or health? Come do an internship with me, @avapamini.bsky.social, Alex Lu, @lcrawford.bsky.social, or Kristen Severson at MSRNE!
Applications are due Dec 1: make sure you include a research statement!
jobs.careers.microsoft.com/global/en/jo...
Applications are due Dec 1: make sure you include a research statement!
jobs.careers.microsoft.com/global/en/jo...
Search Jobs | Microsoft Careers
jobs.careers.microsoft.com
October 21, 2025 at 7:32 PM
Are you a PhD student interested in ML and biology or health? Come do an internship with me, @avapamini.bsky.social, Alex Lu, @lcrawford.bsky.social, or Kristen Severson at MSRNE!
Applications are due Dec 1: make sure you include a research statement!
jobs.careers.microsoft.com/global/en/jo...
Applications are due Dec 1: make sure you include a research statement!
jobs.careers.microsoft.com/global/en/jo...
Reposted by Kevin K. Yang 楊凱筌
Alternate between predicting structure with AF3 and designing sequence with ProteinMPNN to generate proteins, including protein, small molecule and nucleic acid binders
@yehlincho.bsky.social @sokrypton.org
www.biorxiv.org/content/10.1...
@yehlincho.bsky.social @sokrypton.org
www.biorxiv.org/content/10.1...



October 15, 2025 at 10:01 PM
Alternate between predicting structure with AF3 and designing sequence with ProteinMPNN to generate proteins, including protein, small molecule and nucleic acid binders
@yehlincho.bsky.social @sokrypton.org
www.biorxiv.org/content/10.1...
@yehlincho.bsky.social @sokrypton.org
www.biorxiv.org/content/10.1...
Alternate between predicting structure with AF3 and designing sequence with ProteinMPNN to generate proteins, including protein, small molecule and nucleic acid binders
@yehlincho.bsky.social @sokrypton.org
www.biorxiv.org/content/10.1...
@yehlincho.bsky.social @sokrypton.org
www.biorxiv.org/content/10.1...



October 15, 2025 at 10:01 PM
Alternate between predicting structure with AF3 and designing sequence with ProteinMPNN to generate proteins, including protein, small molecule and nucleic acid binders
@yehlincho.bsky.social @sokrypton.org
www.biorxiv.org/content/10.1...
@yehlincho.bsky.social @sokrypton.org
www.biorxiv.org/content/10.1...
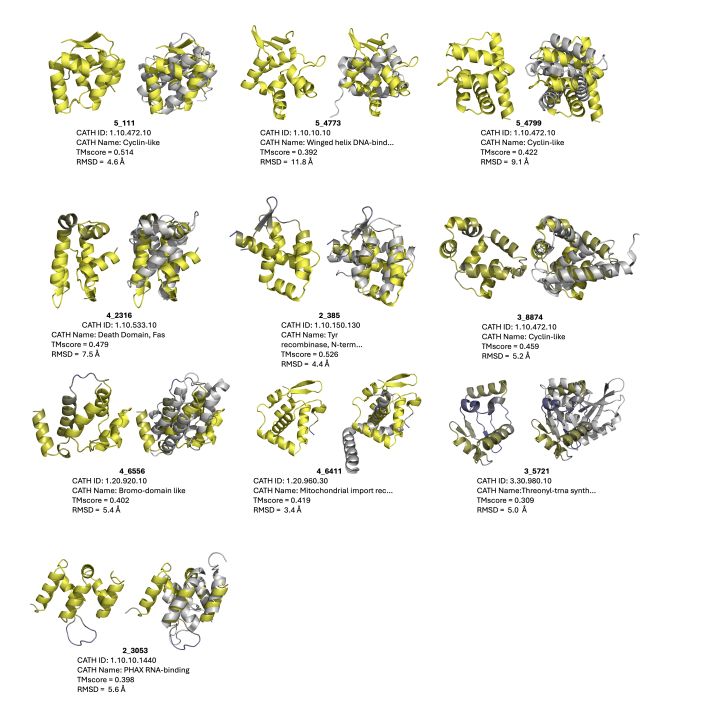
Protein language models can be finetuned to generate many novel structural folds
Arjuna Subramanian, Matt Thomson
www.biorxiv.org/content/10.1...
Arjuna Subramanian, Matt Thomson
www.biorxiv.org/content/10.1...


October 13, 2025 at 2:10 PM
Protein language models can be finetuned to generate many novel structural folds
Arjuna Subramanian, Matt Thomson
www.biorxiv.org/content/10.1...
Arjuna Subramanian, Matt Thomson
www.biorxiv.org/content/10.1...
Reposted by Kevin K. Yang 楊凱筌
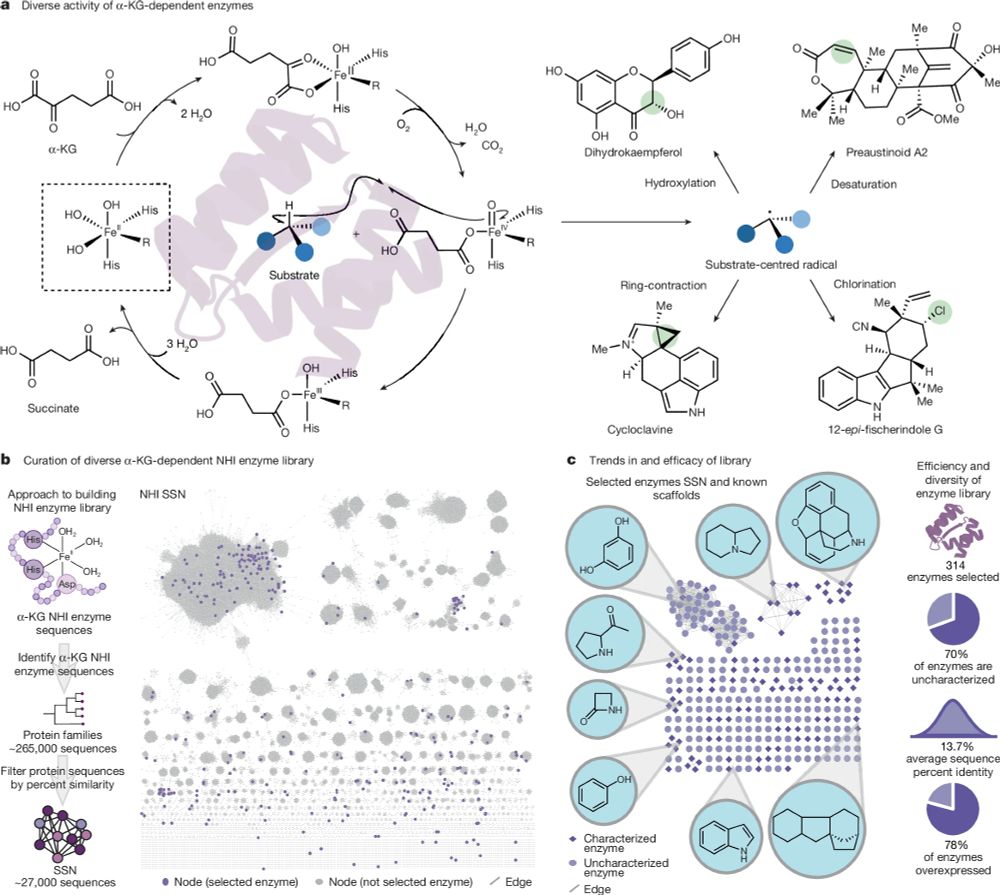
Test the activity of 300+ natural enzymes against 100+ substrates, discover 200+ new enzymatic reactions, and train machine learning models to predict which enzymes can do which reactions.
@aepaton.bsky.social @gabegomes.bsky.social @alisonnarayan.bsky.social
www.nature.com/articles/s41...
@aepaton.bsky.social @gabegomes.bsky.social @alisonnarayan.bsky.social
www.nature.com/articles/s41...

October 2, 2025 at 8:11 PM
Test the activity of 300+ natural enzymes against 100+ substrates, discover 200+ new enzymatic reactions, and train machine learning models to predict which enzymes can do which reactions.
@aepaton.bsky.social @gabegomes.bsky.social @alisonnarayan.bsky.social
www.nature.com/articles/s41...
@aepaton.bsky.social @gabegomes.bsky.social @alisonnarayan.bsky.social
www.nature.com/articles/s41...
Test the activity of 300+ natural enzymes against 100+ substrates, discover 200+ new enzymatic reactions, and train machine learning models to predict which enzymes can do which reactions.
@aepaton.bsky.social @gabegomes.bsky.social @alisonnarayan.bsky.social
www.nature.com/articles/s41...
@aepaton.bsky.social @gabegomes.bsky.social @alisonnarayan.bsky.social
www.nature.com/articles/s41...


October 2, 2025 at 8:11 PM
Test the activity of 300+ natural enzymes against 100+ substrates, discover 200+ new enzymatic reactions, and train machine learning models to predict which enzymes can do which reactions.
@aepaton.bsky.social @gabegomes.bsky.social @alisonnarayan.bsky.social
www.nature.com/articles/s41...
@aepaton.bsky.social @gabegomes.bsky.social @alisonnarayan.bsky.social
www.nature.com/articles/s41...
Combine multimer structure prediction and an antibody language model to design de novo antibodies with nanomolar binding affinity.
@synbiogaolab.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...
@synbiogaolab.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...



September 25, 2025 at 8:02 PM
Combine multimer structure prediction and an antibody language model to design de novo antibodies with nanomolar binding affinity.
@synbiogaolab.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...
@synbiogaolab.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...
Reposted by Kevin K. Yang 楊凱筌
Out of 7 papers in my NeurIPS Benchmarks and Datasets Track area, the PCs overruled my recommendation on 3?!
September 18, 2025 at 12:44 PM
Out of 7 papers in my NeurIPS Benchmarks and Datasets Track area, the PCs overruled my recommendation on 3?!
Reposted by Kevin K. Yang 楊凱筌
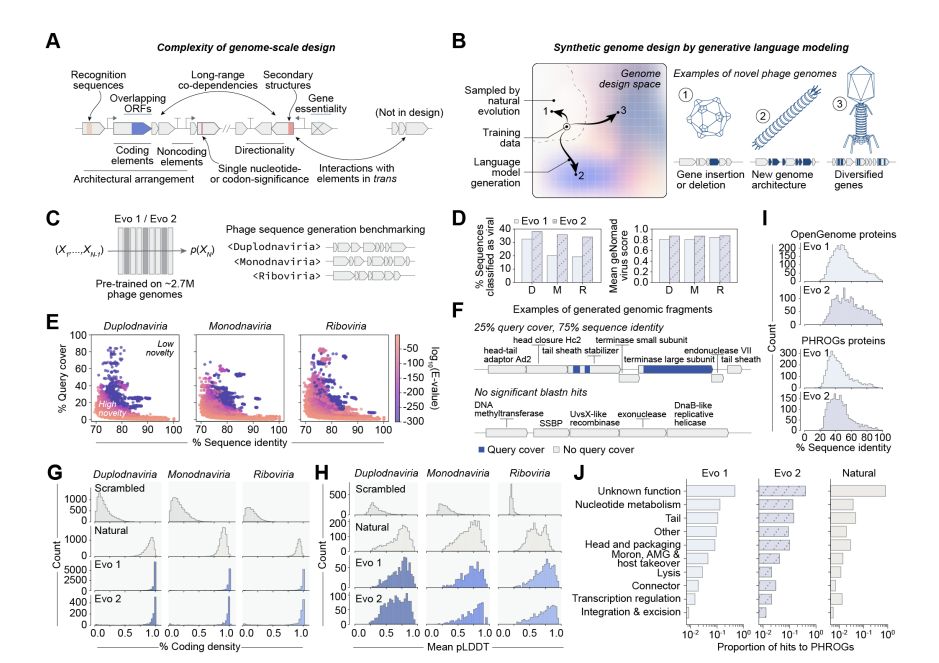
Genome language models can generate new, high-fitness bacteriophages!
@samuelhking.bsky.social @claudiadriscoll.bsky.social
@david-li.bsky.social @danguo.bsky.social @adititm.bsky.social Garyk Brixi @maxewilkinson.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...
@samuelhking.bsky.social @claudiadriscoll.bsky.social
@david-li.bsky.social @danguo.bsky.social @adititm.bsky.social Garyk Brixi @maxewilkinson.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...

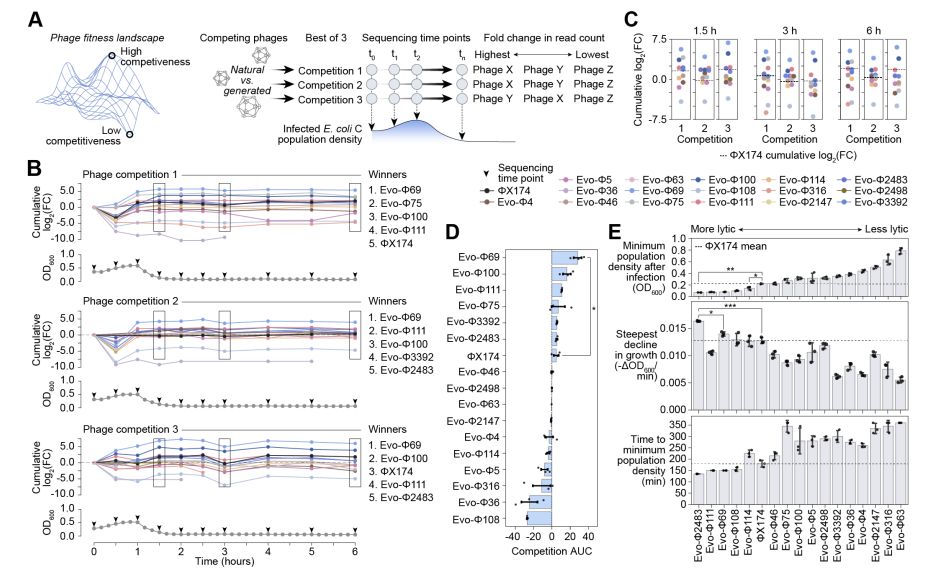

September 17, 2025 at 9:15 PM
Genome language models can generate new, high-fitness bacteriophages!
@samuelhking.bsky.social @claudiadriscoll.bsky.social
@david-li.bsky.social @danguo.bsky.social @adititm.bsky.social Garyk Brixi @maxewilkinson.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...
@samuelhking.bsky.social @claudiadriscoll.bsky.social
@david-li.bsky.social @danguo.bsky.social @adititm.bsky.social Garyk Brixi @maxewilkinson.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...
Out of 7 papers in my NeurIPS Benchmarks and Datasets Track area, the PCs overruled my recommendation on 3?!
September 18, 2025 at 12:44 PM
Out of 7 papers in my NeurIPS Benchmarks and Datasets Track area, the PCs overruled my recommendation on 3?!
Genome language models can generate new, high-fitness bacteriophages!
@samuelhking.bsky.social @claudiadriscoll.bsky.social
@david-li.bsky.social @danguo.bsky.social @adititm.bsky.social Garyk Brixi @maxewilkinson.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...
@samuelhking.bsky.social @claudiadriscoll.bsky.social
@david-li.bsky.social @danguo.bsky.social @adititm.bsky.social Garyk Brixi @maxewilkinson.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...




September 17, 2025 at 9:15 PM
Genome language models can generate new, high-fitness bacteriophages!
@samuelhking.bsky.social @claudiadriscoll.bsky.social
@david-li.bsky.social @danguo.bsky.social @adititm.bsky.social Garyk Brixi @maxewilkinson.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...
@samuelhking.bsky.social @claudiadriscoll.bsky.social
@david-li.bsky.social @danguo.bsky.social @adititm.bsky.social Garyk Brixi @maxewilkinson.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...
The posting is unfortunately not super clear on this, but please apply if you want to do BioML research, especially around machine learning for molecular biology and bioengineering!
I get sad if there are no applicants with this profile in the pool!
I get sad if there are no applicants with this profile in the pool!
If you're an undergrad and want to intern with me, this is where you need to apply!
The Microsoft Research Undergraduate Internship Program offers 12-week internships in our Redmond, NYC, or New England labs for rising juniors and seniors who are passionate about technology. Apply by October 6: msft.it/6015scgSJ
September 15, 2025 at 1:58 PM
The posting is unfortunately not super clear on this, but please apply if you want to do BioML research, especially around machine learning for molecular biology and bioengineering!
I get sad if there are no applicants with this profile in the pool!
I get sad if there are no applicants with this profile in the pool!
If you're an undergrad and want to intern with me, this is where you need to apply!
The Microsoft Research Undergraduate Internship Program offers 12-week internships in our Redmond, NYC, or New England labs for rising juniors and seniors who are passionate about technology. Apply by October 6: msft.it/6015scgSJ

September 13, 2025 at 10:42 AM
If you're an undergrad and want to intern with me, this is where you need to apply!
Train a protein structure predictor that can handle 29 non-canonical amino acids, then use it to design binders with non-canonical amino acids that reduce immunogenicity.
@panhammarstrom.bsky.social @patrickbryant1.bsky.social
www.biorxiv.org/content/10.1...
@panhammarstrom.bsky.social @patrickbryant1.bsky.social
www.biorxiv.org/content/10.1...
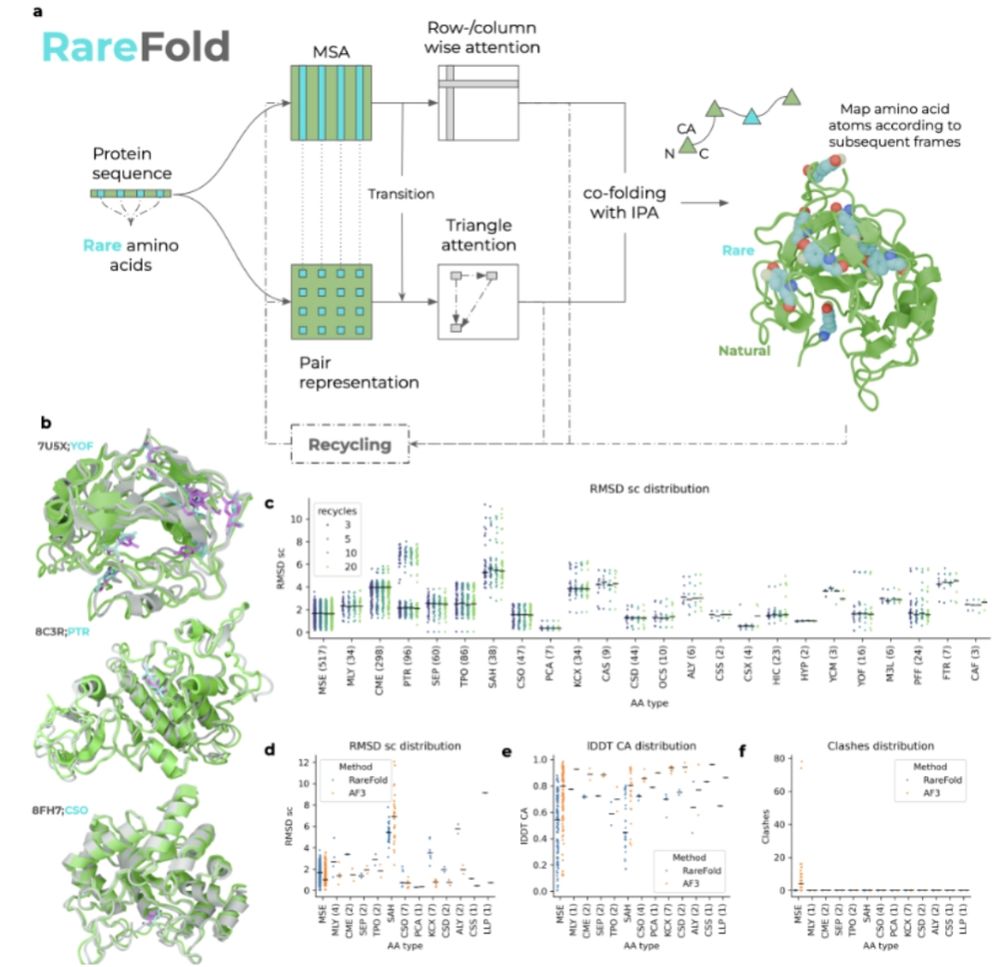

September 9, 2025 at 8:03 PM
Train a protein structure predictor that can handle 29 non-canonical amino acids, then use it to design binders with non-canonical amino acids that reduce immunogenicity.
@panhammarstrom.bsky.social @patrickbryant1.bsky.social
www.biorxiv.org/content/10.1...
@panhammarstrom.bsky.social @patrickbryant1.bsky.social
www.biorxiv.org/content/10.1...
A joint sequence-structure diffusion model for transmembrane proteins!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...


September 5, 2025 at 8:18 PM
A joint sequence-structure diffusion model for transmembrane proteins!
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
A compelling review of how ML/AI could help in the quest to find an enzyme for every reaction.
@jsunn-y.bsky.social @francescazfl.bsky.social Yueming Long @francesarnold.bsky.social
www.cell.com/cell-systems...
@jsunn-y.bsky.social @francescazfl.bsky.social Yueming Long @francesarnold.bsky.social
www.cell.com/cell-systems...

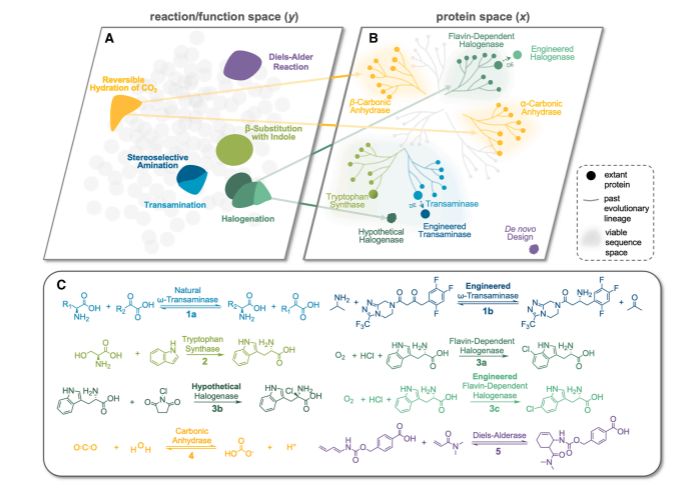
September 3, 2025 at 12:47 PM
A compelling review of how ML/AI could help in the quest to find an enzyme for every reaction.
@jsunn-y.bsky.social @francescazfl.bsky.social Yueming Long @francesarnold.bsky.social
www.cell.com/cell-systems...
@jsunn-y.bsky.social @francescazfl.bsky.social Yueming Long @francesarnold.bsky.social
www.cell.com/cell-systems...
Typical Ohio activity

August 25, 2025 at 12:12 PM
Typical Ohio activity
Reposted by Kevin K. Yang 楊凱筌
Ramblings of a mad AC
1. If you're reviewing a benchmarks and dataset paper, you should look at the dataset and make sure it's not full of garbage.
2. Biological datasets are especially prone to subtle garbage.
1. If you're reviewing a benchmarks and dataset paper, you should look at the dataset and make sure it's not full of garbage.
2. Biological datasets are especially prone to subtle garbage.
August 12, 2025 at 1:43 AM
Ramblings of a mad AC
1. If you're reviewing a benchmarks and dataset paper, you should look at the dataset and make sure it's not full of garbage.
2. Biological datasets are especially prone to subtle garbage.
1. If you're reviewing a benchmarks and dataset paper, you should look at the dataset and make sure it's not full of garbage.
2. Biological datasets are especially prone to subtle garbage.
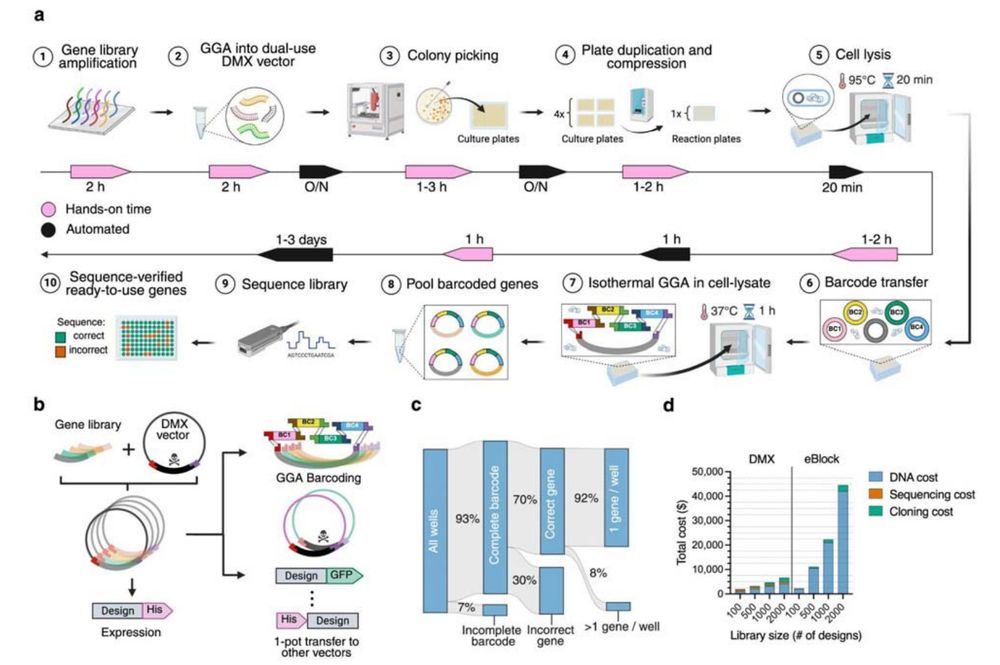
The @uwproteindesign.bsky.social's experimental pipeline behind models like RFdiffusion and ProteinMPNN:
- A rapid, scalable, pipeline for producing and characterizing proteins
- A demultiplexing protocol for converting oligopools to clonal constructs
Jason Qian @lfmilles.bsky.social Basile Wicky
- A rapid, scalable, pipeline for producing and characterizing proteins
- A demultiplexing protocol for converting oligopools to clonal constructs
Jason Qian @lfmilles.bsky.social Basile Wicky
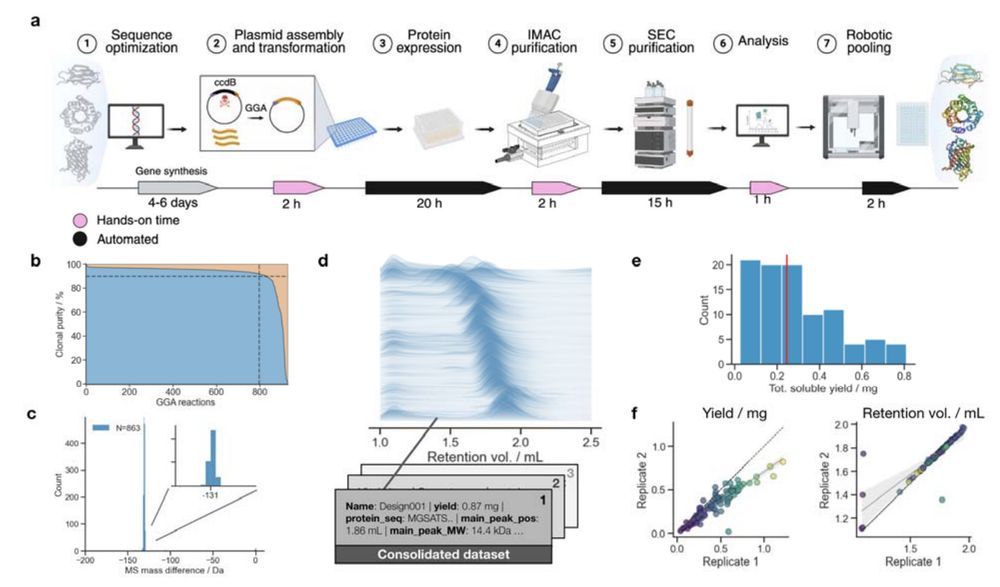
August 12, 2025 at 2:34 PM
The @uwproteindesign.bsky.social's experimental pipeline behind models like RFdiffusion and ProteinMPNN:
- A rapid, scalable, pipeline for producing and characterizing proteins
- A demultiplexing protocol for converting oligopools to clonal constructs
Jason Qian @lfmilles.bsky.social Basile Wicky
- A rapid, scalable, pipeline for producing and characterizing proteins
- A demultiplexing protocol for converting oligopools to clonal constructs
Jason Qian @lfmilles.bsky.social Basile Wicky
Ramblings of a mad AC
1. If you're reviewing a benchmarks and dataset paper, you should look at the dataset and make sure it's not full of garbage.
2. Biological datasets are especially prone to subtle garbage.
1. If you're reviewing a benchmarks and dataset paper, you should look at the dataset and make sure it's not full of garbage.
2. Biological datasets are especially prone to subtle garbage.
August 12, 2025 at 1:43 AM
Ramblings of a mad AC
1. If you're reviewing a benchmarks and dataset paper, you should look at the dataset and make sure it's not full of garbage.
2. Biological datasets are especially prone to subtle garbage.
1. If you're reviewing a benchmarks and dataset paper, you should look at the dataset and make sure it's not full of garbage.
2. Biological datasets are especially prone to subtle garbage.

