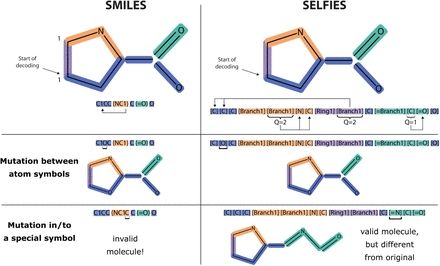
Check the new code: github.com/mqcomplab/bb...

Check the new code: github.com/mqcomplab/bb...
@jsunn-y.bsky.social @francescazfl.bsky.social Yueming Long @francesarnold.bsky.social
www.cell.com/cell-systems...

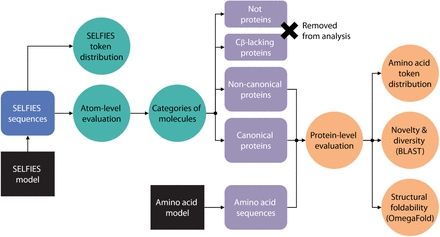
@jsunn-y.bsky.social @francescazfl.bsky.social Yueming Long @francesarnold.bsky.social
www.cell.com/cell-systems...
Our approach allows silencing defense systems of choice. We show how this approach enables programming of “untransformable” bacteria, and how it can enhance phage therapy applications
Congrats Jeremy Garb!
tinyurl.com/Syttt
🧵

Our approach allows silencing defense systems of choice. We show how this approach enables programming of “untransformable” bacteria, and how it can enhance phage therapy applications
Congrats Jeremy Garb!
tinyurl.com/Syttt
🧵
For more details, visit www.ccpbiosim.ac.uk/training2025
The registration link there will open shortly! #compchem

For more details, visit www.ccpbiosim.ac.uk/training2025
The registration link there will open shortly! #compchem
pubs.acs.org/doi/full/10....
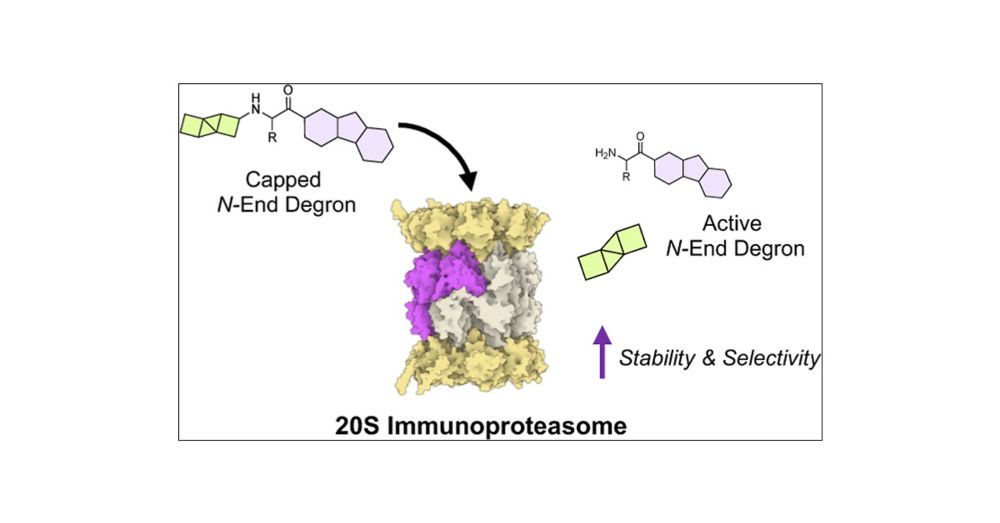
pubs.acs.org/doi/full/10....
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Link: www.nature.com/articles/s42...
[1/4]

Link: www.nature.com/articles/s42...
[1/4]
bit.ly/4lnVYOs

bit.ly/4lnVYOs
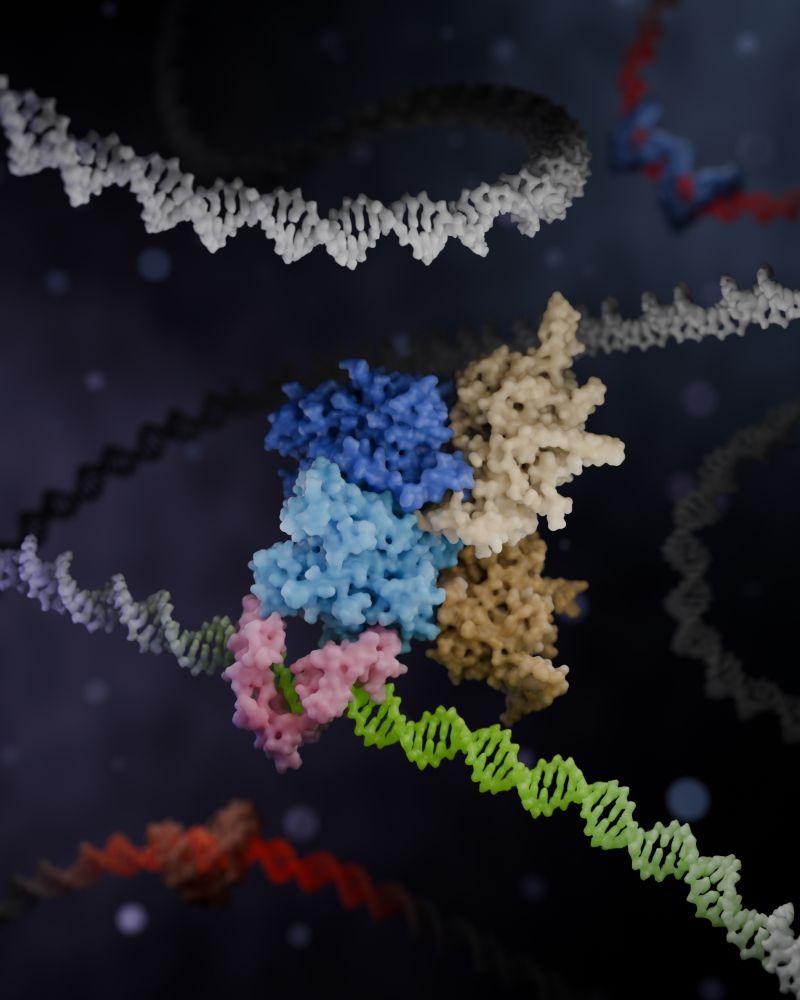
🚨A new coregulator complex, Zincore, acts as a "MOLECULAR GRIP", stabilizing TFs at DNA binding sites across the genome!🧬
💪Proud to be part of this work led by @daaninthelab.bsky.social
Read more: science.org/doi/10.1126/science.adv2861
🚨A new coregulator complex, Zincore, acts as a "MOLECULAR GRIP", stabilizing TFs at DNA binding sites across the genome!🧬
💪Proud to be part of this work led by @daaninthelab.bsky.social
Read more: science.org/doi/10.1126/science.adv2861
🎉Super excited about this milestone as PhD, it’s been a journey, and I’m grateful to finally have these as my first Cryo-EM structures! 🙌
🔁Thank you for sharing!❤️
Read @daaninthelab.bsky.social et. al.: www.science.org/doi/10.1126/...
⬇️ 2nd video
🎉Super excited about this milestone as PhD, it’s been a journey, and I’m grateful to finally have these as my first Cryo-EM structures! 🙌
🔁Thank you for sharing!❤️
Read @daaninthelab.bsky.social et. al.: www.science.org/doi/10.1126/...
⬇️ 2nd video
Cardiolipin inhibits Caspase-4/11!
Huge thanks to my brilliant supervisors Kate & Pablo, to @s-burgener.bsky.social & Mercedes and all the @inflammasomelab.bsky.social, it was a joy working and laughing with you, and to @brozlab.bsky.social & Si Ming lab.

Cardiolipin inhibits Caspase-4/11!
Huge thanks to my brilliant supervisors Kate & Pablo, to @s-burgener.bsky.social & Mercedes and all the @inflammasomelab.bsky.social, it was a joy working and laughing with you, and to @brozlab.bsky.social & Si Ming lab.


The latest @livecomsjournal.bsky.social tutorial by Egger-Hoerschinger et al provides a guide to quantifying hydration thermodynamics using Grid Inhomogeneous Solvation Theory (GIST):
doi.org/10.33011/liv...
#compchem

The latest @livecomsjournal.bsky.social tutorial by Egger-Hoerschinger et al provides a guide to quantifying hydration thermodynamics using Grid Inhomogeneous Solvation Theory (GIST):
doi.org/10.33011/liv...
#compchem

www.nature.com/articles/s41...
www.nature.com/articles/s41...
doi.org/10.33011/liv...
#compchem
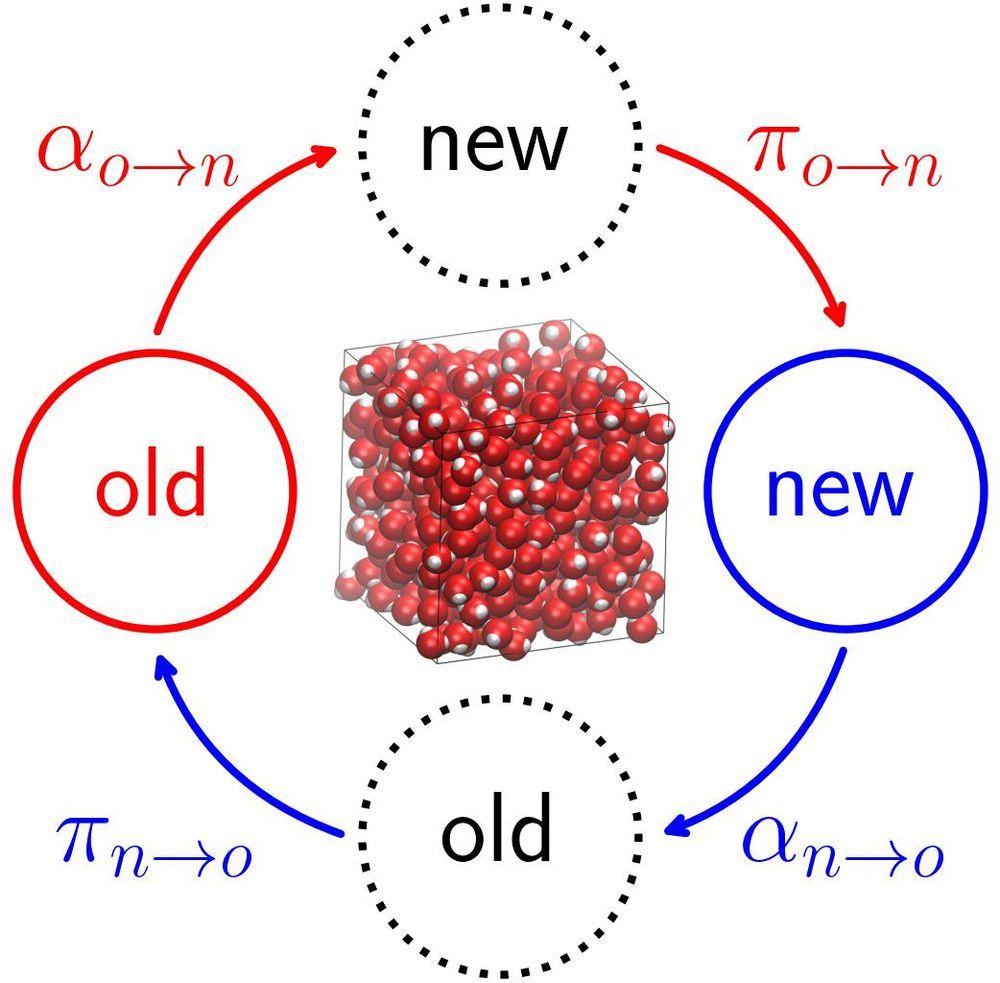
doi.org/10.33011/liv...
#compchem
- deep learning structural metrics only weakly predict success
- The score distribution is different for different types of structures
@grocklin.bsky.social
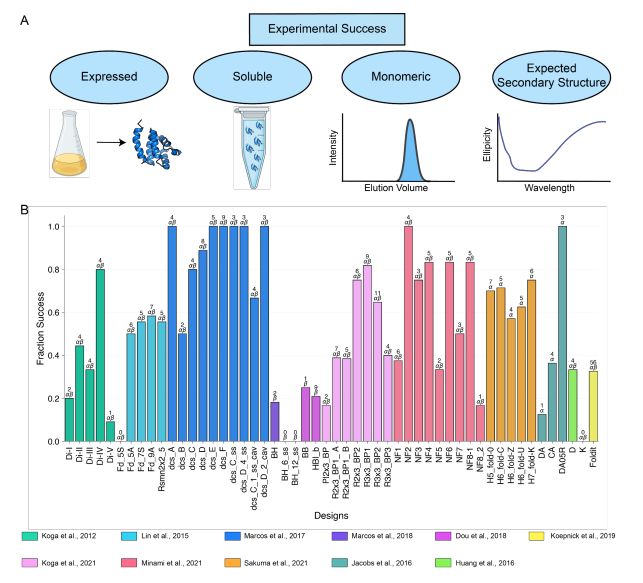



- deep learning structural metrics only weakly predict success
- The score distribution is different for different types of structures
@grocklin.bsky.social
@lynnkamerlin.bsky.social
www.nature.com/articles/s41...
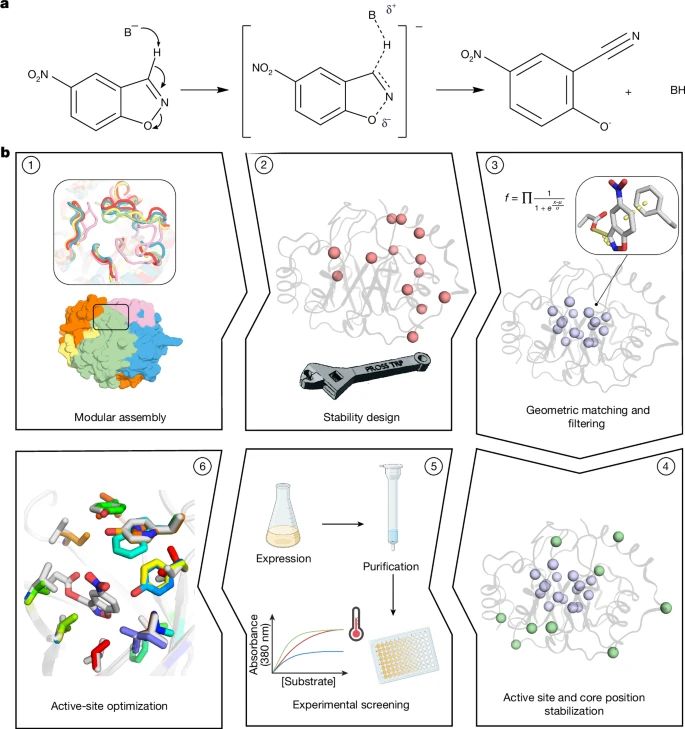
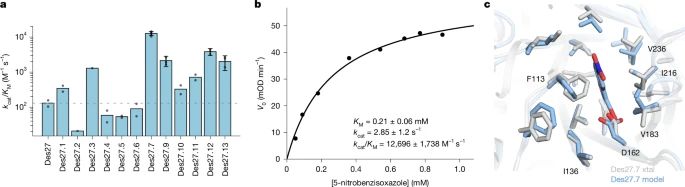
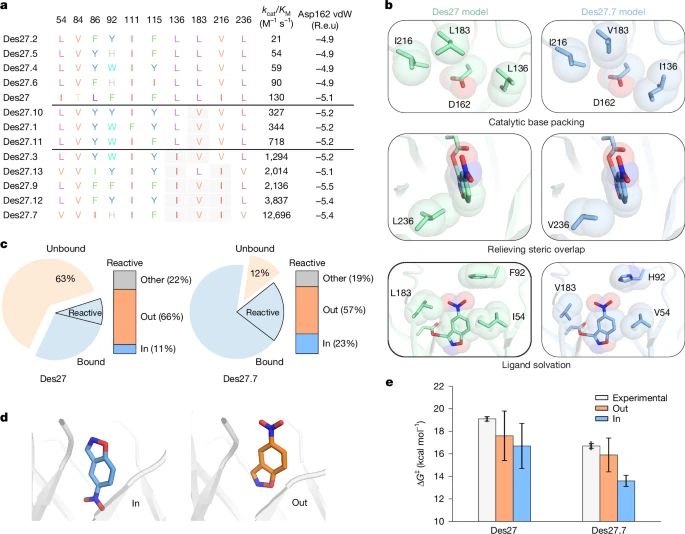
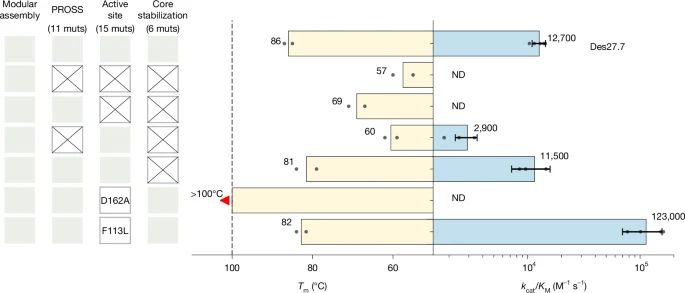
@lynnkamerlin.bsky.social
www.nature.com/articles/s41...