Kieran Didi
@kdidi.bsky.social
🧪 Research Scientist @nvidia and PhD student @Oxford staring at proteins all-day
🧑💻 Website/Blog: https://kdidi.netlify.app/
🤖 GitHub: https://github.com/kierandidi
📚 Prev. Cambridge/Heidelberg
🧑💻 Website/Blog: https://kdidi.netlify.app/
🤖 GitHub: https://github.com/kierandidi
📚 Prev. Cambridge/Heidelberg
Reposted by Kieran Didi
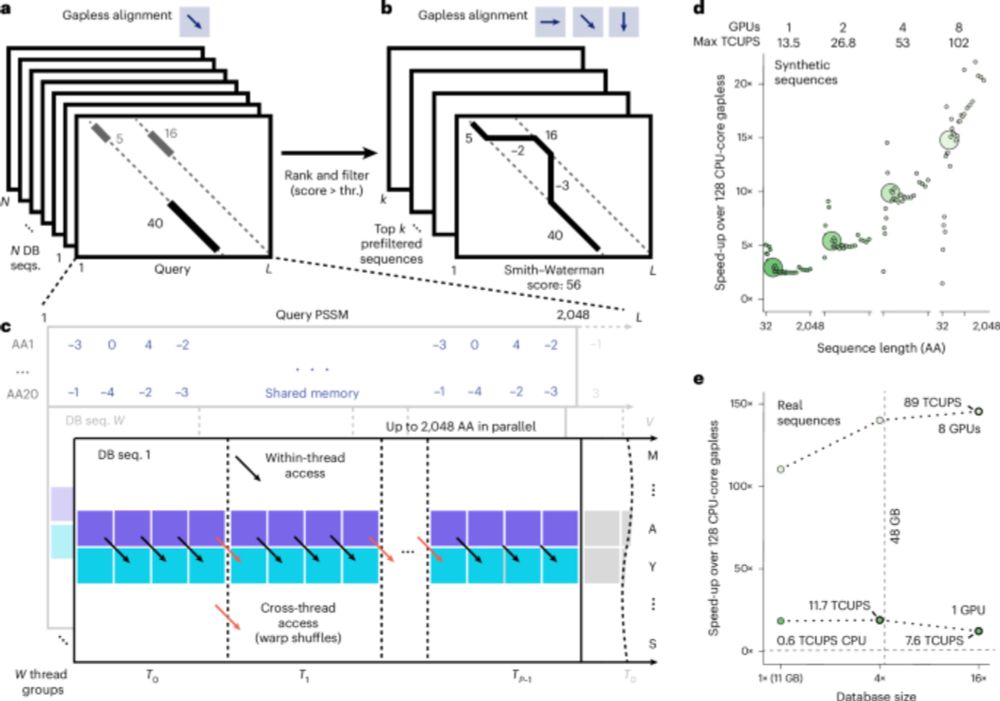
GPU-accelerated MMseqs2 offers tremendous speedup for homology retrieval, protein structure prediction with ColabFold, and protein structure search with Foldseek. @martinsteinegger.bsky.social @milot.bsky.social @machine.learning.bio
www.nature.com/articles/s41...
www.nature.com/articles/s41...
GPU-accelerated homology search with MMseqs2 - Nature Methods
Graphics processing unit-accelerated MMseqs2 offers tremendous speedups for homology retrieval from metagenomic databases, query-centered multiple sequence alignment generation for structure predictio...
www.nature.com
September 18, 2025 at 8:09 PM
GPU-accelerated MMseqs2 offers tremendous speedup for homology retrieval, protein structure prediction with ColabFold, and protein structure search with Foldseek. @martinsteinegger.bsky.social @milot.bsky.social @machine.learning.bio
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Reposted by Kieran Didi
MMseqs2-GPU sets new standards in single query search speed, allows near instant search of big databases, scales to multiple GPUs and is fast beyond VRAM. It enables ColabFold MSA generation in seconds and sub-second Foldseek search against AFDB50. 1/n
📄 www.nature.com/articles/s41...
💿 mmseqs.com
📄 www.nature.com/articles/s41...
💿 mmseqs.com

GPU-accelerated homology search with MMseqs2 - Nature Methods
Graphics processing unit-accelerated MMseqs2 offers tremendous speedups for homology retrieval from metagenomic databases, query-centered multiple sequence alignment generation for structure predictio...
www.nature.com
September 21, 2025 at 8:06 AM
MMseqs2-GPU sets new standards in single query search speed, allows near instant search of big databases, scales to multiple GPUs and is fast beyond VRAM. It enables ColabFold MSA generation in seconds and sub-second Foldseek search against AFDB50. 1/n
📄 www.nature.com/articles/s41...
💿 mmseqs.com
📄 www.nature.com/articles/s41...
💿 mmseqs.com
AtomWorks is out! Building upon @biotite_python, we built a toolkit for all things biomolecules and trained RF3 with it. All open-source, test it via `pip install atomworks`!
AtomWorks: github.com/RosettaCommo...
RF3: github.com/RosettaCommo...
Paper: tinyurl.com/y2w4z65b
1/6
AtomWorks: github.com/RosettaCommo...
RF3: github.com/RosettaCommo...
Paper: tinyurl.com/y2w4z65b
1/6

August 15, 2025 at 5:44 PM
AtomWorks is out! Building upon @biotite_python, we built a toolkit for all things biomolecules and trained RF3 with it. All open-source, test it via `pip install atomworks`!
AtomWorks: github.com/RosettaCommo...
RF3: github.com/RosettaCommo...
Paper: tinyurl.com/y2w4z65b
1/6
AtomWorks: github.com/RosettaCommo...
RF3: github.com/RosettaCommo...
Paper: tinyurl.com/y2w4z65b
1/6
Reposted by Kieran Didi
RosettaFold 3 is here! 🧬🚀
AtomWorks (the foundational data pipeline powering it) is perhaps the really most exciting part of this release!
Congratulations @simonmathis.bsky.social and team!!! ❤️
bioRxiv preprint: www.biorxiv.org/content/10.1...
AtomWorks (the foundational data pipeline powering it) is perhaps the really most exciting part of this release!
Congratulations @simonmathis.bsky.social and team!!! ❤️
bioRxiv preprint: www.biorxiv.org/content/10.1...

August 15, 2025 at 1:26 PM
RosettaFold 3 is here! 🧬🚀
AtomWorks (the foundational data pipeline powering it) is perhaps the really most exciting part of this release!
Congratulations @simonmathis.bsky.social and team!!! ❤️
bioRxiv preprint: www.biorxiv.org/content/10.1...
AtomWorks (the foundational data pipeline powering it) is perhaps the really most exciting part of this release!
Congratulations @simonmathis.bsky.social and team!!! ❤️
bioRxiv preprint: www.biorxiv.org/content/10.1...
Reposted by Kieran Didi
(1/7)
Training biomolecular foundation models shouldn't be so hard. And open-source structure prediction is important. So today we're releasing two software packages: AtomWorks and RosettaFold3 (RF3)
[https://www.biorxiv.org/content/10.1101/2025.08.14.670328v2](www.biorxiv.org/content/10.1...)
Training biomolecular foundation models shouldn't be so hard. And open-source structure prediction is important. So today we're releasing two software packages: AtomWorks and RosettaFold3 (RF3)
[https://www.biorxiv.org/content/10.1101/2025.08.14.670328v2](www.biorxiv.org/content/10.1...)

Accelerating Biomolecular Modeling with AtomWorks and RF3
Deep learning methods trained on protein structure databases have revolutionized biomolecular structure prediction, but developing and training new models remains a considerable challenge. To facilita...
www.biorxiv.org
August 15, 2025 at 5:17 PM
(1/7)
Training biomolecular foundation models shouldn't be so hard. And open-source structure prediction is important. So today we're releasing two software packages: AtomWorks and RosettaFold3 (RF3)
[https://www.biorxiv.org/content/10.1101/2025.08.14.670328v2](www.biorxiv.org/content/10.1...)
Training biomolecular foundation models shouldn't be so hard. And open-source structure prediction is important. So today we're releasing two software packages: AtomWorks and RosettaFold3 (RF3)
[https://www.biorxiv.org/content/10.1101/2025.08.14.670328v2](www.biorxiv.org/content/10.1...)
Very excited about our latest all-atom generative model proteina, check out the project page (research.nvidia.com/labs/genair/...) and stay tuned for the code release soon!
July 19, 2025 at 8:46 PM
Very excited about our latest all-atom generative model proteina, check out the project page (research.nvidia.com/labs/genair/...) and stay tuned for the code release soon!
Excited to present my first paper officially as a PhD student now as an ICLR Oral this week! Super fun work with the GenAIR team at NVIDIA.
Talk: Fr 10:54 - 11:06 (Oral Session 3B, Garnet 213-215)
Poster: Fr 15:00-17:30 (Hall 3 + Hall 2B #5)
Come by the poster/reach out to chat
Talk: Fr 10:54 - 11:06 (Oral Session 3B, Garnet 213-215)
Poster: Fr 15:00-17:30 (Hall 3 + Hall 2B #5)
Come by the poster/reach out to chat
April 23, 2025 at 8:37 AM
Excited to present my first paper officially as a PhD student now as an ICLR Oral this week! Super fun work with the GenAIR team at NVIDIA.
Talk: Fr 10:54 - 11:06 (Oral Session 3B, Garnet 213-215)
Poster: Fr 15:00-17:30 (Hall 3 + Hall 2B #5)
Come by the poster/reach out to chat
Talk: Fr 10:54 - 11:06 (Oral Session 3B, Garnet 213-215)
Poster: Fr 15:00-17:30 (Hall 3 + Hall 2B #5)
Come by the poster/reach out to chat
Such a fun project to work on with a stellar team! Stay tuned for other things to come here, and see you all in Singapore!
📢📢 "Proteina: Scaling Flow-based Protein Structure Generative Models"
#ICLR2025 (Oral Presentation)
🔥 Project page: research.nvidia.com/labs/genair/...
📜 Paper: arxiv.org/abs/2503.00710
🛠️ Code and weights: github.com/NVIDIA-Digit...
🧵Details in thread...
(1/n)
#ICLR2025 (Oral Presentation)
🔥 Project page: research.nvidia.com/labs/genair/...
📜 Paper: arxiv.org/abs/2503.00710
🛠️ Code and weights: github.com/NVIDIA-Digit...
🧵Details in thread...
(1/n)
March 4, 2025 at 5:44 PM
Such a fun project to work on with a stellar team! Stay tuned for other things to come here, and see you all in Singapore!
Reposted by Kieran Didi
Yet another story of issues with benchmarks and evaluations in ML4bio + a much stronger and fair benchmark #bioMLeval
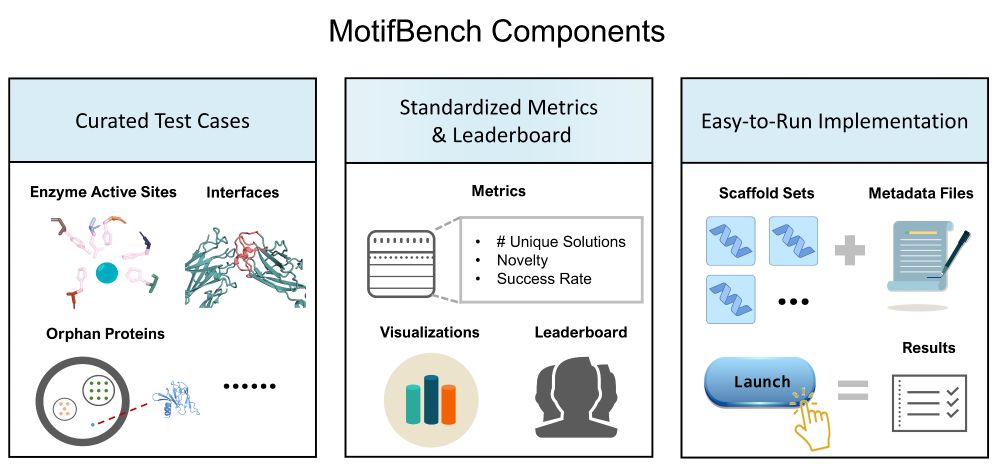
🔥 Benchmark Alert! MotifBench sets a new standard for evaluating protein design methods in motif scaffolding.
Why does this matter? Reproducibility & fair comparison have been lacking—until now.
Paper: arxiv.org/abs/2502.12479 | Repo: github.com/blt2114/Moti...
A thread ⬇️
Why does this matter? Reproducibility & fair comparison have been lacking—until now.
Paper: arxiv.org/abs/2502.12479 | Repo: github.com/blt2114/Moti...
A thread ⬇️
February 20, 2025 at 6:00 AM
Yet another story of issues with benchmarks and evaluations in ML4bio + a much stronger and fair benchmark #bioMLeval
Have a look at our shiny new benchmark for motif-scaffolding in computational protein design! New (and harder) tasks, including a reproducible evaluation pipeline
🔥 Benchmark Alert! MotifBench sets a new standard for evaluating protein design methods in motif scaffolding.
Why does this matter? Reproducibility & fair comparison have been lacking—until now.
Paper: arxiv.org/abs/2502.12479 | Repo: github.com/blt2114/Moti...
A thread ⬇️
Why does this matter? Reproducibility & fair comparison have been lacking—until now.
Paper: arxiv.org/abs/2502.12479 | Repo: github.com/blt2114/Moti...
A thread ⬇️

February 20, 2025 at 3:51 PM
Have a look at our shiny new benchmark for motif-scaffolding in computational protein design! New (and harder) tasks, including a reproducible evaluation pipeline
Reposted by Kieran Didi
This!
Also well put in this editorial in PLOS Comp Biol:
Putting benchmarks in their rightful place: The heart of computational biology
doi.org/10.1371/jour...
Also well put in this editorial in PLOS Comp Biol:
Putting benchmarks in their rightful place: The heart of computational biology
doi.org/10.1371/jour...

January 15, 2025 at 12:08 PM
This!
Also well put in this editorial in PLOS Comp Biol:
Putting benchmarks in their rightful place: The heart of computational biology
doi.org/10.1371/jour...
Also well put in this editorial in PLOS Comp Biol:
Putting benchmarks in their rightful place: The heart of computational biology
doi.org/10.1371/jour...
Reposted by Kieran Didi
Our paper on computational design of chemically induced protein interactions is out in @natureportfolio.bsky.social. Big thanks to all co-authors, especially Anthony Marchand, Stephen Buckley and Bruno Correia!
t.co/vtYlhi8aQm
t.co/vtYlhi8aQm
January 15, 2025 at 4:37 PM
Our paper on computational design of chemically induced protein interactions is out in @natureportfolio.bsky.social. Big thanks to all co-authors, especially Anthony Marchand, Stephen Buckley and Bruno Correia!
t.co/vtYlhi8aQm
t.co/vtYlhi8aQm
Reposted by Kieran Didi
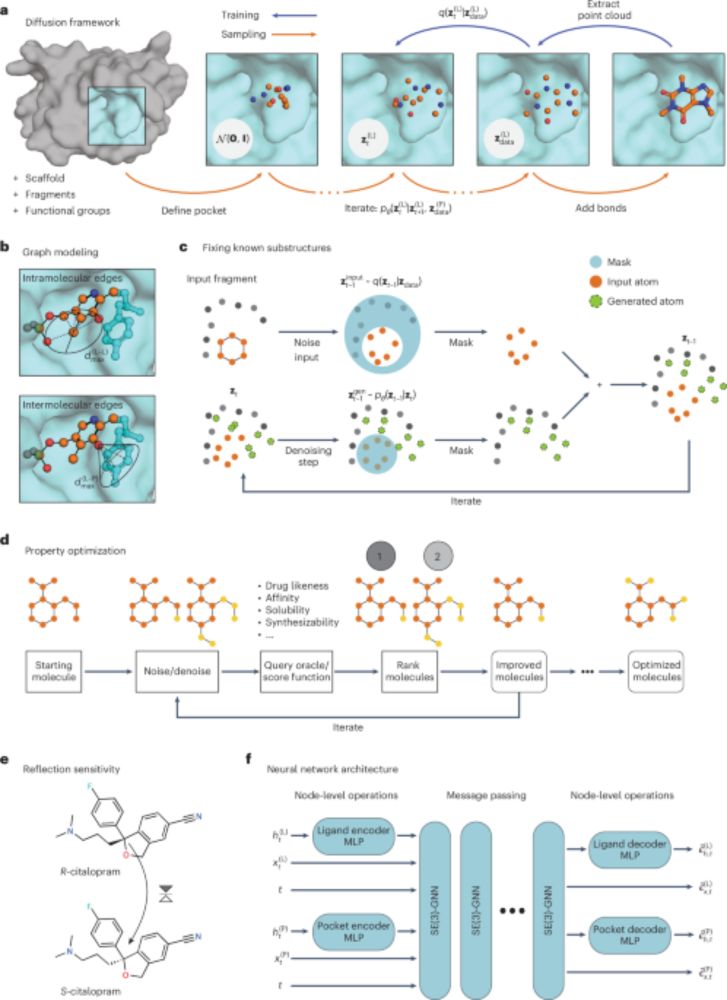
After two years, our paper on generative models for structure-based drug design is finally out in @natcomputsci.bsky.social
www.nature.com/articles/s43...
www.nature.com/articles/s43...
Structure-based drug design with equivariant diffusion models - Nature Computational Science
This work applies diffusion models to conditional molecule generation and shows how they can be used to tackle various structure-based drug design problems
www.nature.com
December 9, 2024 at 2:00 PM
After two years, our paper on generative models for structure-based drug design is finally out in @natcomputsci.bsky.social
www.nature.com/articles/s43...
www.nature.com/articles/s43...
Reposted by Kieran Didi
Thanks Jascha 🫶
We’re working hard to create sustainable funding mechanisms for open source scientific tooling - understanding the challenge landscape is a key first step!
We’re working hard to create sustainable funding mechanisms for open source scientific tooling - understanding the challenge landscape is a key first step!
Rory @rory.bio is working on a large project to accelerate science 🧪 through high-quality & open-source software.
To steer the project it would be amazing to hear from scientists across fields about problems in the scientific process *you* want to see solved! Tell Rory here flywhl-ideas.notion.site
To steer the project it would be amazing to hear from scientists across fields about problems in the scientific process *you* want to see solved! Tell Rory here flywhl-ideas.notion.site

Notion – The all-in-one workspace for your notes, tasks, wikis, and databases.
A new tool that blends your everyday work apps into one. It's the all-in-one workspace for you and your team
flywhl-ideas.notion.site
December 3, 2024 at 10:45 AM
Thanks Jascha 🫶
We’re working hard to create sustainable funding mechanisms for open source scientific tooling - understanding the challenge landscape is a key first step!
We’re working hard to create sustainable funding mechanisms for open source scientific tooling - understanding the challenge landscape is a key first step!
Love PyMOL Remote, one of these tools that does one thing and does it well!
For some more guidance on how to use this, Martin Buttenschön wrote a nice blogpost: www.blopig.com/blog/2024/11...
November 29, 2024 at 11:28 AM
Love PyMOL Remote, one of these tools that does one thing and does it well!
Reposted by Kieran Didi
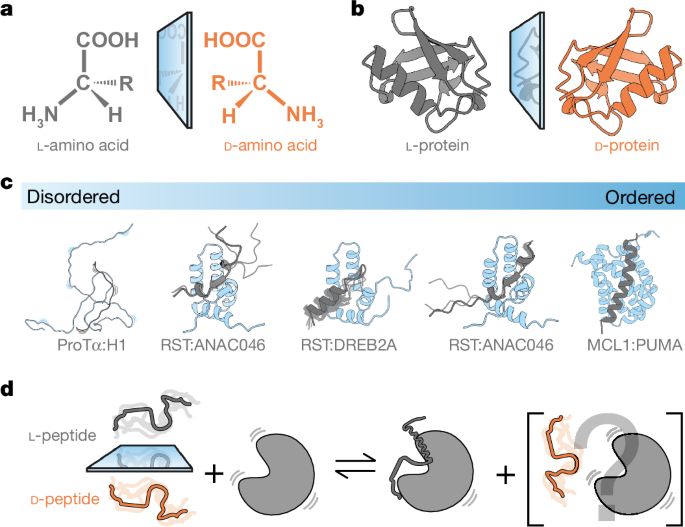
This should go chiral
We did this crazy project where we tried to see if proteins could interact with their mirror image ligand. Seems impossible when proteins need to form 3D structures to interact. But what about if the interaction remains disordered???
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Stereochemistry in the disorder–order continuum of protein interactions - Nature
Studies on protein–protein interactions using proteins containing d- or l-amino acids show that stereoselectivity of binding varies with the degree of disorder within the complex.
www.nature.com
November 27, 2024 at 8:35 PM
This should go chiral
MSAs go brrr with MMseqs2-GPU! Super fun project, happy to work with and learn from a stellar team of engineers and scientists. Try it out and stay tuned!
📄 Preprint: www.biorxiv.org/content/10.1...
💾 Code: mmseqs.com
🗞️ Blog: developer.nvidia.com/blog/boost-a...
📄 Preprint: www.biorxiv.org/content/10.1...
💾 Code: mmseqs.com
🗞️ Blog: developer.nvidia.com/blog/boost-a...
November 16, 2024 at 2:18 PM
MSAs go brrr with MMseqs2-GPU! Super fun project, happy to work with and learn from a stellar team of engineers and scientists. Try it out and stay tuned!
📄 Preprint: www.biorxiv.org/content/10.1...
💾 Code: mmseqs.com
🗞️ Blog: developer.nvidia.com/blog/boost-a...
📄 Preprint: www.biorxiv.org/content/10.1...
💾 Code: mmseqs.com
🗞️ Blog: developer.nvidia.com/blog/boost-a...

