
PhD student at the University of Cambridge
Prev. at FAIR, Prescient Design, and MRC LMB
📝 https://chaitjo.substack.com

📝: tinyurl.com/gRNAde-paper
Unlike proteins, RNA design has long relied on "wisdom of the crowd" (human experts) or the slow crawl of directed evolution — gRNAde changes that! 🧵👇
Happy to have played a small part in the new RNAPro model, significantly outperforming AlphaFold 3 as well as VFold (CASP winners)

Happy to have played a small part in the new RNAPro model, significantly outperforming AlphaFold 3 as well as VFold (CASP winners)
Sharing something interesting: Friso van de Stadt created a pretty neat explainer video for our gRNAde paper and "AlphaGo moment for RNA design" blogpost!
www.youtube.com/watch?v=wDeZ...

Sharing something interesting: Friso van de Stadt created a pretty neat explainer video for our gRNAde paper and "AlphaGo moment for RNA design" blogpost!
www.youtube.com/watch?v=wDeZ...
💌: chaitjo.substack.com/p/an-ai-rese...

💌: chaitjo.substack.com/p/an-ai-rese...
I pride myself on open-science & this is probably the most intense release I've done!

I pride myself on open-science & this is probably the most intense release I've done!
It provides standardised metrics for validity, stability, & much more. Already includes results for 12 models!
🔗 Paper: arxiv.org/abs/2512.04562
1/4

It provides standardised metrics for validity, stability, & much more. Already includes results for 12 models!
🔗 Paper: arxiv.org/abs/2512.04562
1/4
Over 5,000 participants from 114 countries competed to build AI models that predict cellular responses to genetic perturbations. Today we're announcing the winners and reflecting on what we learned.

Over 5,000 participants from 114 countries competed to build AI models that predict cellular responses to genetic perturbations. Today we're announcing the winners and reflecting on what we learned.
→ becoming a mainstream term in academia soon, in few years (Overton window)
→ becoming a mainstream term in academia soon, in few years (Overton window)
The polymerase ribozyme results are pretty cool too. 😎
📝: tinyurl.com/gRNAde-paper
Unlike proteins, RNA design has long relied on "wisdom of the crowd" (human experts) or the slow crawl of directed evolution — gRNAde changes that! 🧵👇

The polymerase ribozyme results are pretty cool too. 😎
📝: tinyurl.com/gRNAde-paper
Unlike proteins, RNA design has long relied on "wisdom of the crowd" (human experts) or the slow crawl of directed evolution — gRNAde changes that! 🧵👇

📝: tinyurl.com/gRNAde-paper
Unlike proteins, RNA design has long relied on "wisdom of the crowd" (human experts) or the slow crawl of directed evolution — gRNAde changes that! 🧵👇

📝: tinyurl.com/gRNAde-paper
Unlike proteins, RNA design has long relied on "wisdom of the crowd" (human experts) or the slow crawl of directed evolution — gRNAde changes that! 🧵👇
Generative inverse design of RNA structure and function with gRNAde
www.biorxiv.org/content/10.1...

pmc.ncbi.nlm.nih.gov/articles/PMC...
pmc.ncbi.nlm.nih.gov/articles/PMC...
Its an important moment for structure-based biomolecule design: models starting to work and action shifting from academia to industry.
So what are the next scientific problems academia could be thinking about?
chaitjo.substack.com/p/beyond-str...

Its an important moment for structure-based biomolecule design: models starting to work and action shifting from academia to industry.
So what are the next scientific problems academia could be thinking about?
chaitjo.substack.com/p/beyond-str...
I think its because training deep learning models is less like science/engineering, and more like cooking. It takes some time to develop the intuitions around learning dynamics of big models.
I think its because training deep learning models is less like science/engineering, and more like cooking. It takes some time to develop the intuitions around learning dynamics of big models.

Find information on paper guidelines at mlsb.io. Submissions will be made through CMT.
Find information on paper guidelines at mlsb.io. Submissions will be made through CMT.
@samuelhking.bsky.social @claudiadriscoll.bsky.social
@david-li.bsky.social @danguo.bsky.social @adititm.bsky.social Garyk Brixi @maxewilkinson.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...


@samuelhking.bsky.social @claudiadriscoll.bsky.social
@david-li.bsky.social @danguo.bsky.social @adititm.bsky.social Garyk Brixi @maxewilkinson.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...
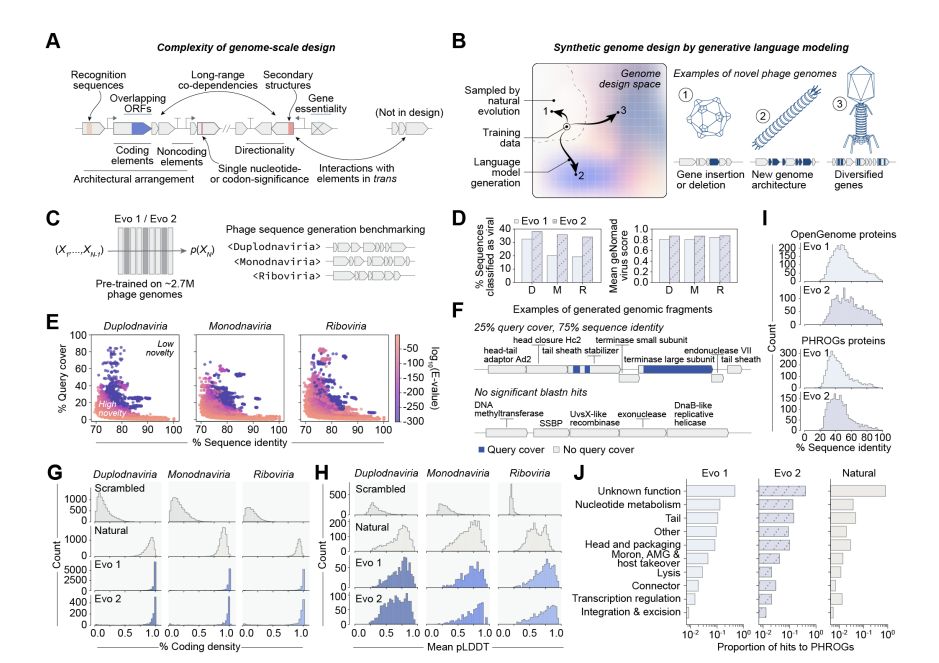
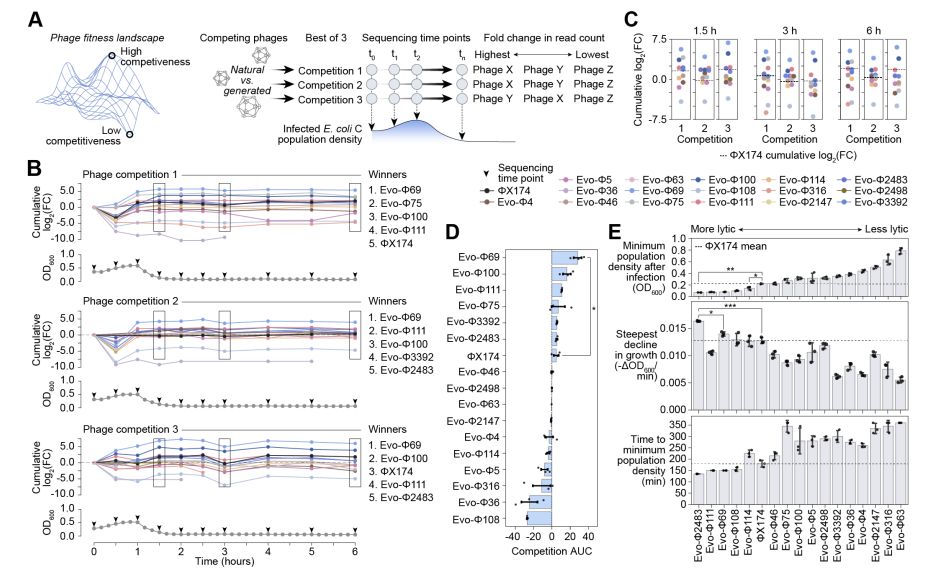
Today, we share our preprint “Generative design of novel bacteriophages with genome language models”, where we validate the first, functional AI-generated genomes 🧵
Today, we share our preprint “Generative design of novel bacteriophages with genome language models”, where we validate the first, functional AI-generated genomes 🧵
with generative bioML, i'm always looking at how similar the generated sequences are to known sequences. let's take a look
Today, we share our preprint “Generative design of novel bacteriophages with genome language models”, where we validate the first, functional AI-generated genomes 🧵
with generative bioML, i'm always looking at how similar the generated sequences are to known sequences. let's take a look
Two workshops (San Diego & Copenhagen) will run concurrently to support broader attendance. You can indicate your location preference(s) in the submission portal💫

Two workshops (San Diego & Copenhagen) will run concurrently to support broader attendance. You can indicate your location preference(s) in the submission portal💫
(Slide from Denny Zhou's Stanford talk on LLM Reasoning)

(Slide from Denny Zhou's Stanford talk on LLM Reasoning)

Training biomolecular foundation models shouldn't be so hard. And open-source structure prediction is important. So today we're releasing two software packages: AtomWorks and RosettaFold3 (RF3)
[https://www.biorxiv.org/content/10.1101/2025.08.14.670328v2](www.biorxiv.org/content/10.1...)

Training biomolecular foundation models shouldn't be so hard. And open-source structure prediction is important. So today we're releasing two software packages: AtomWorks and RosettaFold3 (RF3)
[https://www.biorxiv.org/content/10.1101/2025.08.14.670328v2](www.biorxiv.org/content/10.1...)
🗓️ Deadline: September 26, 2025 🔗 More info: cmt3.research.microsoft.com/MLSB2025
🗓️ Deadline: September 26, 2025 🔗 More info: cmt3.research.microsoft.com/MLSB2025

