https://chaitjo.com
(Slide from Denny Zhou's Stanford talk on LLM Reasoning)

(Slide from Denny Zhou's Stanford talk on LLM Reasoning)
AtomWorks (the foundational data pipeline powering it) is perhaps the really most exciting part of this release!
Congratulations @simonmathis.bsky.social and team!!! ❤️
bioRxiv preprint: www.biorxiv.org/content/10.1...

AtomWorks (the foundational data pipeline powering it) is perhaps the really most exciting part of this release!
Congratulations @simonmathis.bsky.social and team!!! ❤️
bioRxiv preprint: www.biorxiv.org/content/10.1...
Introducing Dynamic MPNN - our first step towards truly multi-state and 'programmable' protein design! Great collaboration led by Alex Abrudan and Sebastian Pujalte!

Introducing Dynamic MPNN - our first step towards truly multi-state and 'programmable' protein design! Great collaboration led by Alex Abrudan and Sebastian Pujalte!
This is (very) old news, so what's new?

This is (very) old news, so what's new?
Just a thought: Maybe Boltz-2 is also a moment similar to GPT-3 where a sizeable part of the community moves on from academic publishing
and to the world of capabilities, tech reports, and shipping models

Just a thought: Maybe Boltz-2 is also a moment similar to GPT-3 where a sizeable part of the community moves on from academic publishing
and to the world of capabilities, tech reports, and shipping models
Here's a (hopefully) simple repo to instal locally on laptop/single-GPU and a handy notebook to play around with!
github.com/chaitjo/alph...

Here's a (hopefully) simple repo to instal locally on laptop/single-GPU and a handy notebook to play around with!
github.com/chaitjo/alph...
All-atom DiT with minimal molecular inductive bias is on par with SOTA equivariant diffusion (SemlaFlow, a very strong and optimized model)
Scaling pure Transformers can alleviate explicit bond prediction!

All-atom DiT with minimal molecular inductive bias is on par with SOTA equivariant diffusion (SemlaFlow, a very strong and optimized model)
Scaling pure Transformers can alleviate explicit bond prediction!
@pranam.bsky.social @kishwar.bsky.social @bunnech.bsky.social and many others!

@pranam.bsky.social @kishwar.bsky.social @bunnech.bsky.social and many others!
So excited that the ML world will gather in Singapore - And hoping to help people have a great time here! 🤗




So excited that the ML world will gather in Singapore - And hoping to help people have a great time here! 🤗
- briefly summarises the big ideas and key takeaways
Link - www.chaitjo.com/publication/...

- briefly summarises the big ideas and key takeaways
Link - www.chaitjo.com/publication/...
I'll be speaking about All Atom Diffusion Transformers for the first time publicly!
Thank you @erikjbekkers.bsky.social for the invitation 🙏
www.nsbm.nl

I'll be speaking about All Atom Diffusion Transformers for the first time publicly!
Thank you @erikjbekkers.bsky.social for the invitation 🙏
www.nsbm.nl
I've fixed a lot of small issues and tested on two different GPU clusters to ensure everything should now work out of the box.
PS. lightning-hydra template makes for very modular code :)

I've fixed a lot of small issues and tested on two different GPU clusters to ensure everything should now work out of the box.
PS. lightning-hydra template makes for very modular code :)
**Modelling microbiome-mediated epigenetic inheritance of disease risk**
💻 Calendar invite, virtual link, and all the details: talks.cam.ac.uk/talk/index/2...

**Modelling microbiome-mediated epigenetic inheritance of disease risk**
💻 Calendar invite, virtual link, and all the details: talks.cam.ac.uk/talk/index/2...
There’s huge excitement for universal interatomic ML potentials rn — we’re trying to think about the equivalent for universal generative foundation models and inverse molecular design!

There’s huge excitement for universal interatomic ML potentials rn — we’re trying to think about the equivalent for universal generative foundation models and inverse molecular design!
A. We don’t necessarily need SO3 equivariance for generative models, if we scale data/denoiser/compute
B. Similar for math-y discrete + continuous diffusion, as unified generative modelling in latent space is simple

A. We don’t necessarily need SO3 equivariance for generative models, if we scale data/denoiser/compute
B. Similar for math-y discrete + continuous diffusion, as unified generative modelling in latent space is simple
Latent diffusion transformers can sample new, realistic latents -- which we decode to valid molecules/crystals.
ADiT can be told whether to sample a molecule or crystal using classifier-free guidance with class tokens (+ other properties possible)

Latent diffusion transformers can sample new, realistic latents -- which we decode to valid molecules/crystals.
ADiT can be told whether to sample a molecule or crystal using classifier-free guidance with class tokens (+ other properties possible)
We can embed molecules and materials to the same latent embedding space, where we hope to learn shared representations of atomic interactions.
Its simple to train a joint autoencoder for all-atom reconstruction of molecules, crystals, and beyond.

We can embed molecules and materials to the same latent embedding space, where we hope to learn shared representations of atomic interactions.
Its simple to train a joint autoencoder for all-atom reconstruction of molecules, crystals, and beyond.
An amazing collaboration w/ Xiang Fu, Yi-Lun Liao, Vahe Garekhanyan, Ben Miller, Anuroop Sriram and Zack Ulissi 👊

An amazing collaboration w/ Xiang Fu, Yi-Lun Liao, Vahe Garekhanyan, Ben Miller, Anuroop Sriram and Zack Ulissi 👊
— towards Foundation Models for generative chemistry, from my internship with the FAIR Chemistry team
There are a couple ML ideas which I think are new and exciting in here 👇

— towards Foundation Models for generative chemistry, from my internship with the FAIR Chemistry team
There are a couple ML ideas which I think are new and exciting in here 👇
What started as a side project over 2 years ago has lead to a spotlight paper and a new scientific journey for me --
Here I am learning how to pipette :D

What started as a side project over 2 years ago has lead to a spotlight paper and a new scientific journey for me --
Here I am learning how to pipette :D
- Attention across multi-heads and layers can be seen as a heterogenous, dynamically evolving graph.
- Attention graphs are complex systems represent information flow in Transformers.
- We can use network science to extract mechanistic insights from them!

- Attention across multi-heads and layers can be seen as a heterogenous, dynamically evolving graph.
- Attention graphs are complex systems represent information flow in Transformers.
- We can use network science to extract mechanistic insights from them!
A wonderful collaboration with superstar MPhil students Batu El, Deepro Choudhury, as well as Pietro Liò as part of the Geometric Deep Learning class at @cst.cam.ac.uk
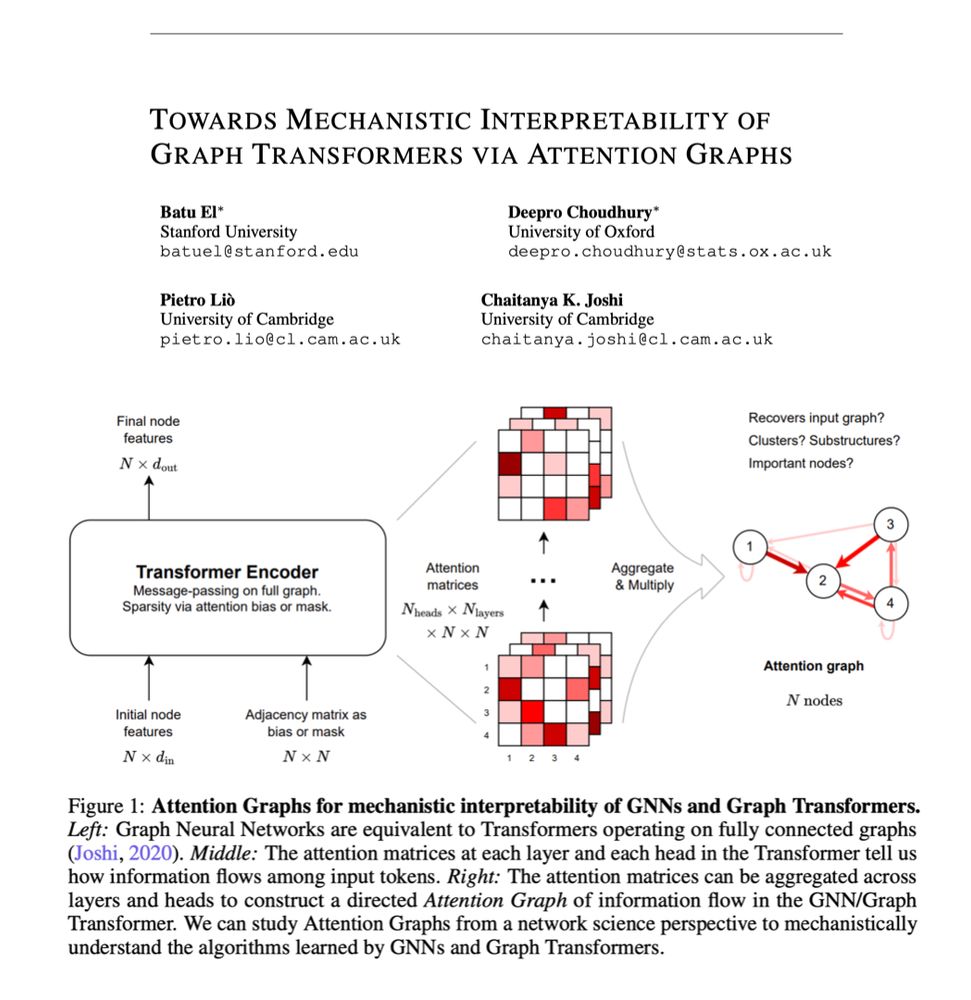
A wonderful collaboration with superstar MPhil students Batu El, Deepro Choudhury, as well as Pietro Liò as part of the Geometric Deep Learning class at @cst.cam.ac.uk

A short journey from JJ Thomson Avenue (electrons) to Francis Crick Avenue (nucleic acids) :)

A short journey from JJ Thomson Avenue (electrons) to Francis Crick Avenue (nucleic acids) :)
— and I must say there is something really addictive about the product sold in UK which is so obviously more tasty than the one in Sg or US. Just by frequency of how much I’m consuming it.

— and I must say there is something really addictive about the product sold in UK which is so obviously more tasty than the one in Sg or US. Just by frequency of how much I’m consuming it.

