Kieran Didi
@kdidi.bsky.social
🧪 Research Scientist @nvidia and PhD student @Oxford staring at proteins all-day
🧑💻 Website/Blog: https://kdidi.netlify.app/
🤖 GitHub: https://github.com/kierandidi
📚 Prev. Cambridge/Heidelberg
🧑💻 Website/Blog: https://kdidi.netlify.app/
🤖 GitHub: https://github.com/kierandidi
📚 Prev. Cambridge/Heidelberg
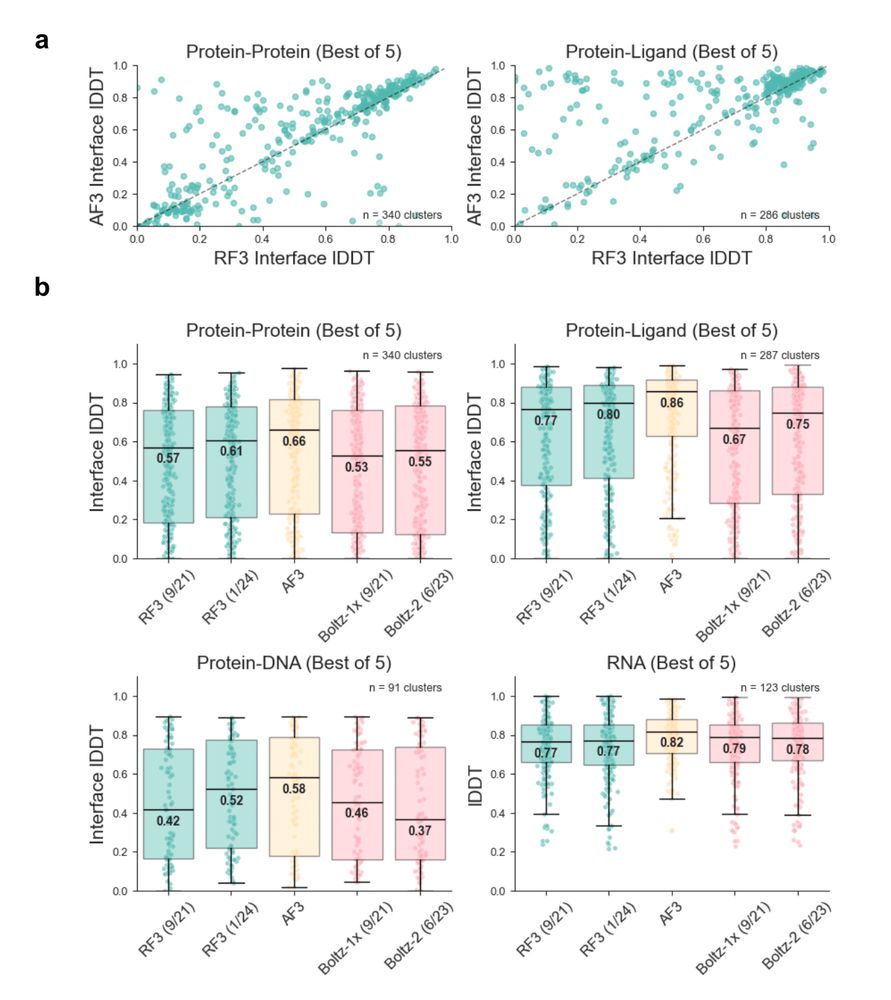
The preprint shows how atomworks leads to better reference conformers (and better predictions!), enables advanced features in RF3 like chirality-aware training or ligand templating and narrows the performance gap to closed-source models. 5/6
August 15, 2025 at 5:47 PM
The preprint shows how atomworks leads to better reference conformers (and better predictions!), enables advanced features in RF3 like chirality-aware training or ligand templating and narrows the performance gap to closed-source models. 5/6
`atomworks.ml` on the other hand offers advanced dataset featurization and sampling for deep learning workflows, all operating on the canonical AtomArray object from @biotite_python so that all transforms are traceable and generalizable between models. 4/6

August 15, 2025 at 5:46 PM
`atomworks.ml` on the other hand offers advanced dataset featurization and sampling for deep learning workflows, all operating on the canonical AtomArray object from @biotite_python so that all transforms are traceable and generalizable between models. 4/6
AtomWorks has two main components: atomworks.io takes a file (cif, sdf, ...) and does parsing, cleaning and more. You can also look at your structures in a notebook or via PyMol thanks to pymol-remote, so you can directly inspect if your code does what you want! 3/6

August 15, 2025 at 5:45 PM
AtomWorks has two main components: atomworks.io takes a file (cif, sdf, ...) and does parsing, cleaning and more. You can also look at your structures in a notebook or via PyMol thanks to pymol-remote, so you can directly inspect if your code does what you want! 3/6
In the past, every BioML model had its own data pipeline, creating loads of overhead. With AtomWorks, >80% of code is shared between models like ProteinMPNN, RF3 or design models. 2/6

August 15, 2025 at 5:45 PM
In the past, every BioML model had its own data pipeline, creating loads of overhead. With AtomWorks, >80% of code is shared between models like ProteinMPNN, RF3 or design models. 2/6
AtomWorks is out! Building upon @biotite_python, we built a toolkit for all things biomolecules and trained RF3 with it. All open-source, test it via `pip install atomworks`!
AtomWorks: github.com/RosettaCommo...
RF3: github.com/RosettaCommo...
Paper: tinyurl.com/y2w4z65b
1/6
AtomWorks: github.com/RosettaCommo...
RF3: github.com/RosettaCommo...
Paper: tinyurl.com/y2w4z65b
1/6

August 15, 2025 at 5:44 PM
AtomWorks is out! Building upon @biotite_python, we built a toolkit for all things biomolecules and trained RF3 with it. All open-source, test it via `pip install atomworks`!
AtomWorks: github.com/RosettaCommo...
RF3: github.com/RosettaCommo...
Paper: tinyurl.com/y2w4z65b
1/6
AtomWorks: github.com/RosettaCommo...
RF3: github.com/RosettaCommo...
Paper: tinyurl.com/y2w4z65b
1/6
Excited to present my first paper officially as a PhD student now as an ICLR Oral this week! Super fun work with the GenAIR team at NVIDIA.
Talk: Fr 10:54 - 11:06 (Oral Session 3B, Garnet 213-215)
Poster: Fr 15:00-17:30 (Hall 3 + Hall 2B #5)
Come by the poster/reach out to chat
Talk: Fr 10:54 - 11:06 (Oral Session 3B, Garnet 213-215)
Poster: Fr 15:00-17:30 (Hall 3 + Hall 2B #5)
Come by the poster/reach out to chat
April 23, 2025 at 8:37 AM
Excited to present my first paper officially as a PhD student now as an ICLR Oral this week! Super fun work with the GenAIR team at NVIDIA.
Talk: Fr 10:54 - 11:06 (Oral Session 3B, Garnet 213-215)
Poster: Fr 15:00-17:30 (Hall 3 + Hall 2B #5)
Come by the poster/reach out to chat
Talk: Fr 10:54 - 11:06 (Oral Session 3B, Garnet 213-215)
Poster: Fr 15:00-17:30 (Hall 3 + Hall 2B #5)
Come by the poster/reach out to chat

