
ML & computational biology 🤖🧬⚛️
📚 Paper: openreview.net/forum?id=g3V...
💻 Code: github.com/LPDI-EPFL/Dr...
(1/4)

Two workshops (San Diego & Copenhagen) will run concurrently to support broader attendance. You can indicate your location preference(s) in the submission portal💫

Two workshops (San Diego & Copenhagen) will run concurrently to support broader attendance. You can indicate your location preference(s) in the submission portal💫
go.nature.com/46bhOAv

go.nature.com/46bhOAv
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Registration is now open at cemm.at/aithyra-symp...

Registration is now open at cemm.at/aithyra-symp...
What is BioEmu? Check out this video:
youtu.be/LStKhWcL0VE?...
What is BioEmu? Check out this video:
youtu.be/LStKhWcL0VE?...
www.science.org/doi/10.1126/...
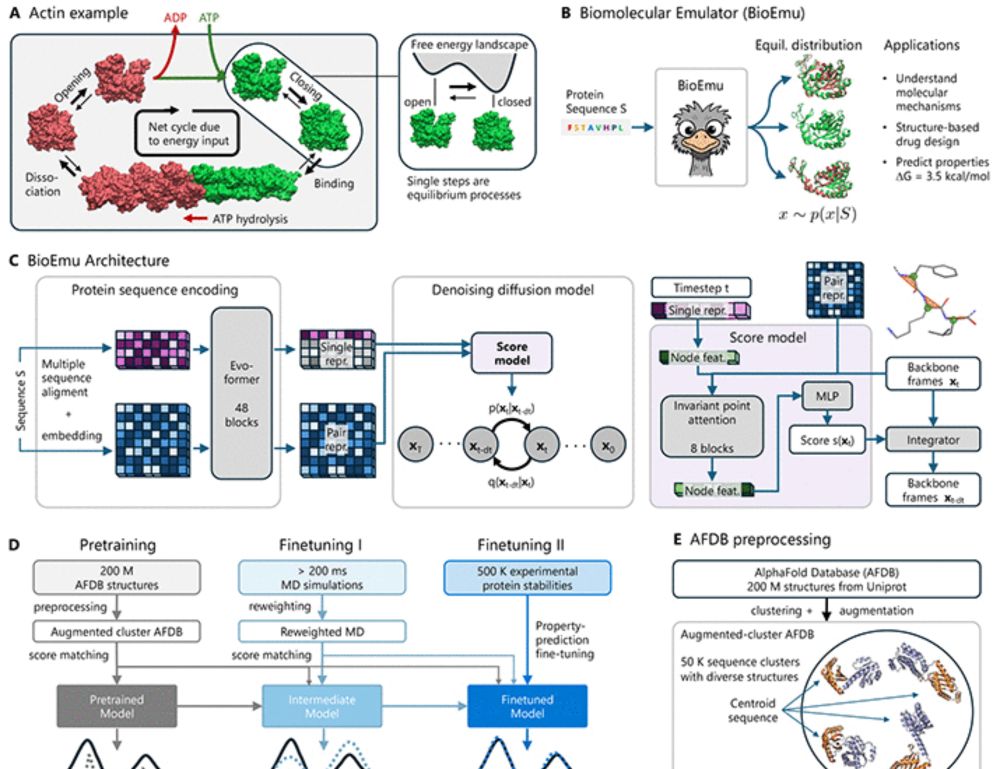
www.science.org/doi/10.1126/...
Learn more in this week's issue: scim.ag/44uRorm

Learn more in this week's issue: scim.ag/44uRorm
github.com/martinpacesa...
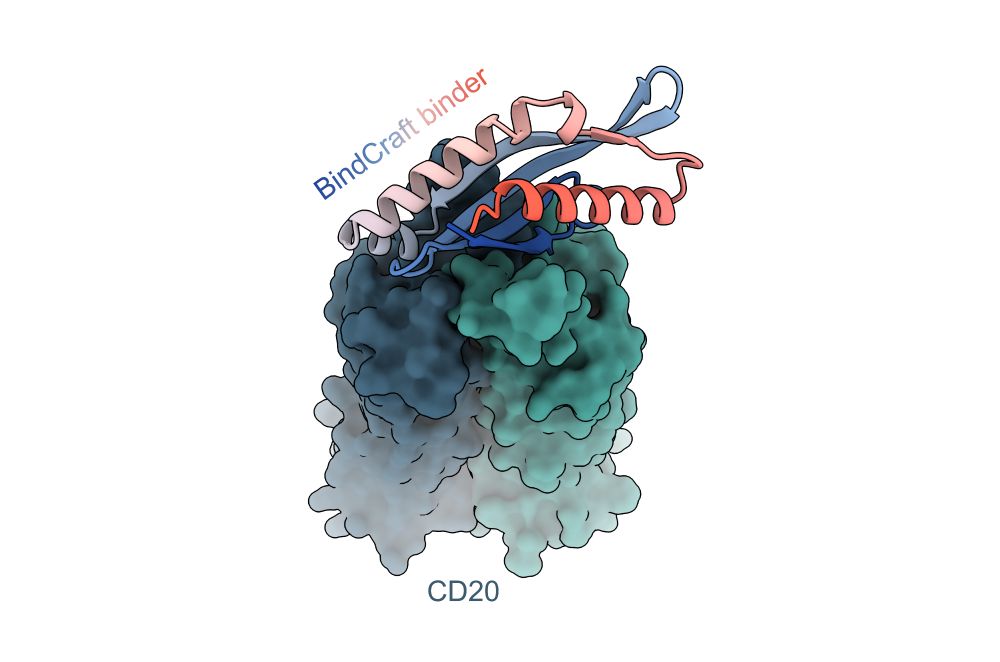
github.com/martinpacesa...
📃 openreview.net/forum?id=bZW...
🔗 github.com/rneeser/Late...
A thread 🧵👇

📃 openreview.net/forum?id=bZW...
🔗 github.com/rneeser/Late...
A thread 🧵👇
🔗 doi.org/10.1101/2025...
💻 github.com/Bitbol-Lab/r...
1/7

🔗 doi.org/10.1101/2025...
💻 github.com/Bitbol-Lab/r...
1/7
Thanks @jjimenezluna.bsky.social for the help!
🌐 colab.research.google.com/github/sokry...
📄 www.biorxiv.org/content/10.1...

Thanks @jjimenezluna.bsky.social for the help!
🌐 colab.research.google.com/github/sokry...
📄 www.biorxiv.org/content/10.1...
We apply synthetic biology and protein design to address key challenges in immunotherapy.
lit.eu/work-with-us/
#SynBio #ProteinDesign #Immunotherapy

We apply synthetic biology and protein design to address key challenges in immunotherapy.
lit.eu/work-with-us/
#SynBio #ProteinDesign #Immunotherapy
Additional details in thread 🧵
#ICLR2025
📚 Paper: openreview.net/forum?id=g3V...
💻 Code: github.com/LPDI-EPFL/Dr...
(1/4)
Additional details in thread 🧵
#ICLR2025
— towards Foundation Models for generative chemistry, from my internship with the FAIR Chemistry team
There are a couple ML ideas which I think are new and exciting in here 👇

— towards Foundation Models for generative chemistry, from my internship with the FAIR Chemistry team
There are a couple ML ideas which I think are new and exciting in here 👇
📚 Paper: openreview.net/forum?id=g3V...
💻 Code: github.com/LPDI-EPFL/Dr...
(1/4)
📚 Paper: openreview.net/forum?id=g3V...
💻 Code: github.com/LPDI-EPFL/Dr...
(1/4)
Please go ahead, play with it and let us know if there are issues.
github.com/microsoft/bi...
Please go ahead, play with it and let us know if there are issues.
github.com/microsoft/bi...

t.co/vtYlhi8aQm
t.co/vtYlhi8aQm
BioEmu from MSR: Scalable emulation of protein equilibrium ensembles with generative deep learning www.biorxiv.org/content/10.1...
Looking forward to hearing more explanainers in tomorrow's reading group session on zoom at 9am PT / 12pm ET / 6pm CET: portal.valencelabs.com/logg
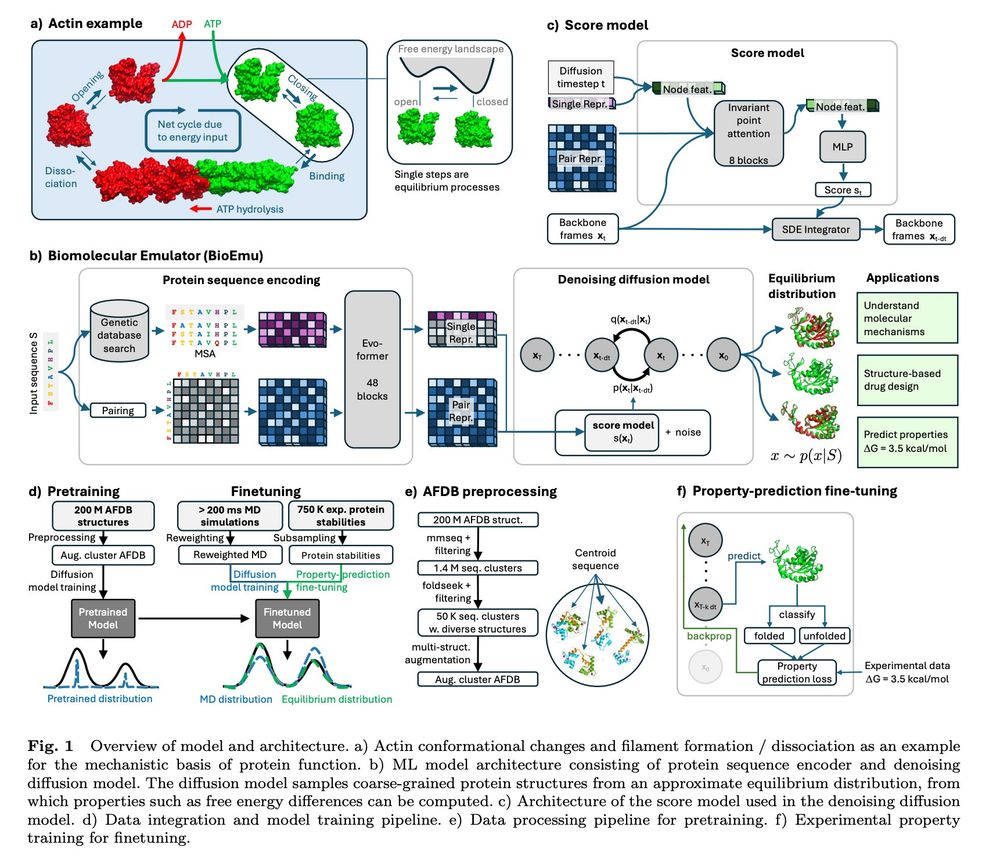
BioEmu from MSR: Scalable emulation of protein equilibrium ensembles with generative deep learning www.biorxiv.org/content/10.1...
Looking forward to hearing more explanainers in tomorrow's reading group session on zoom at 9am PT / 12pm ET / 6pm CET: portal.valencelabs.com/logg
www.nature.com/articles/s43...
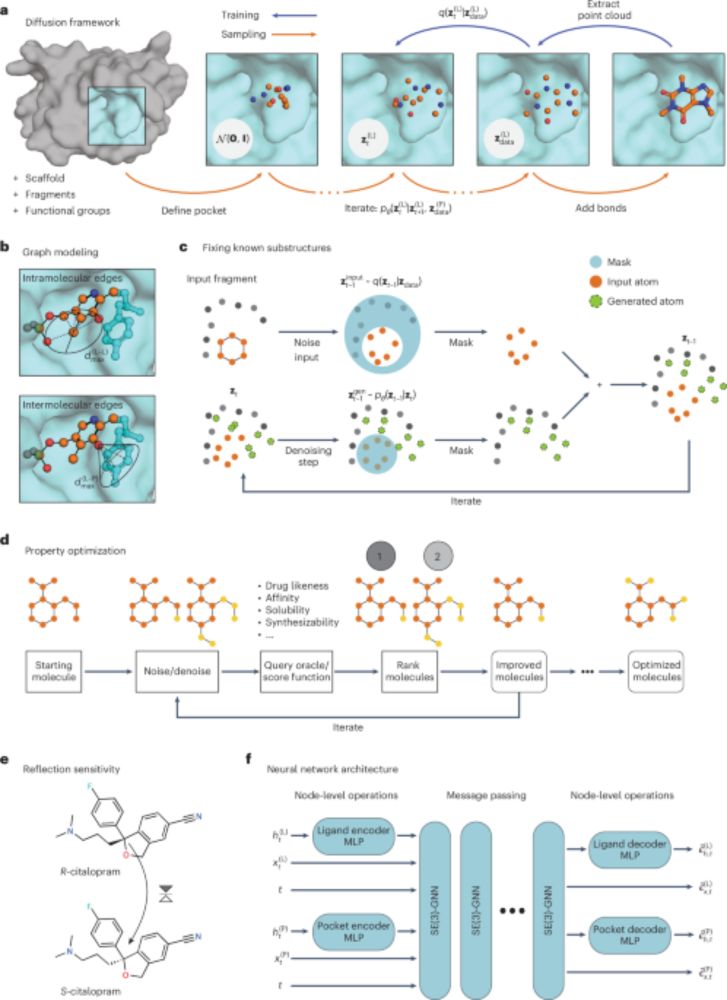
www.nature.com/articles/s43...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...


