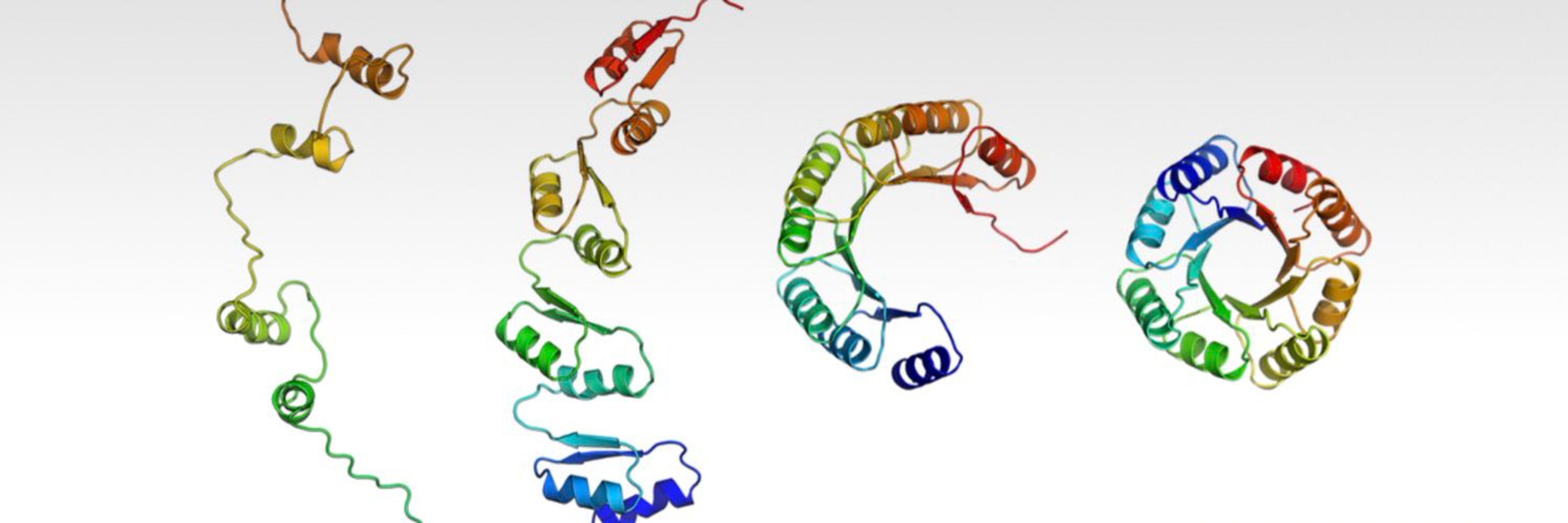
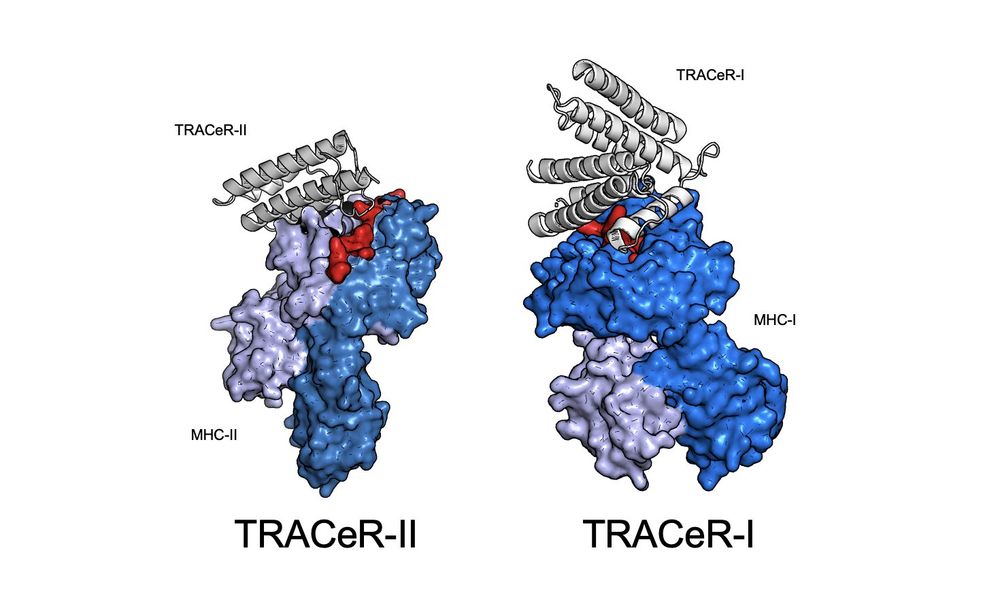
TRACeR for MHC-I: go.nature.com/4gcLzn5
TRACeR for MHC-II: go.nature.com/4gj5OQk

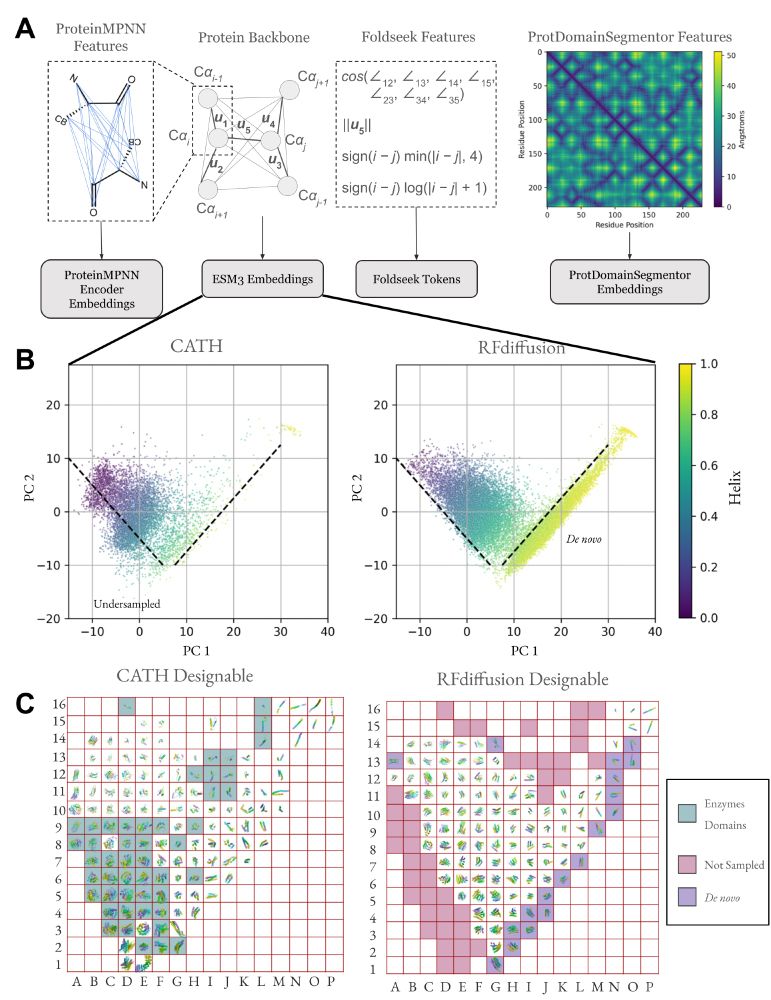
SLAE reasons over atomic interactions to recover structures and residue pairwise energetics, yielding a generalizable, physics-informed latent space. (1/8)

SLAE reasons over atomic interactions to recover structures and residue pairwise energetics, yielding a generalizable, physics-informed latent space. (1/8)
Feedback and requests are welcome!
Feedback and requests are welcome!

@richardshuai.bsky.social Talal Widatalla @possuhuanglab.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...
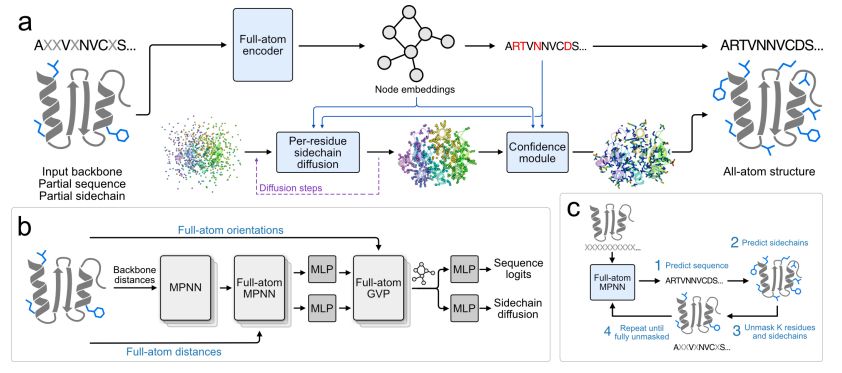
@richardshuai.bsky.social Talal Widatalla @possuhuanglab.bsky.social @brianhie.bsky.social
www.biorxiv.org/content/10.1...
@possuhuanglab.bsky.social
www.biorxiv.org/content/10.1...
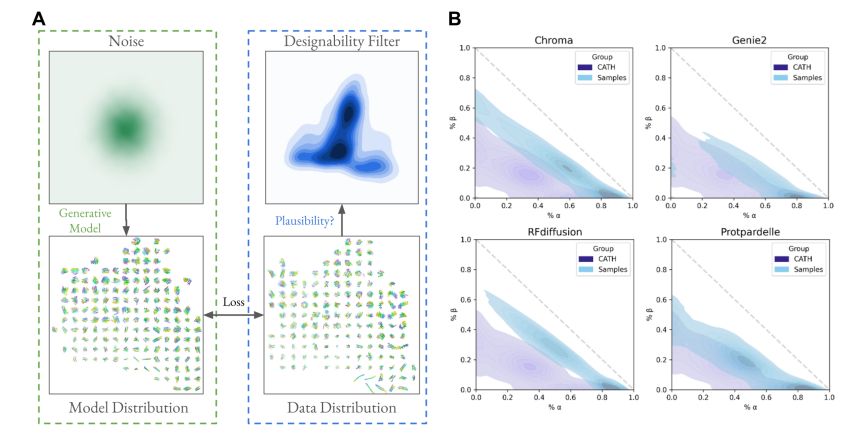
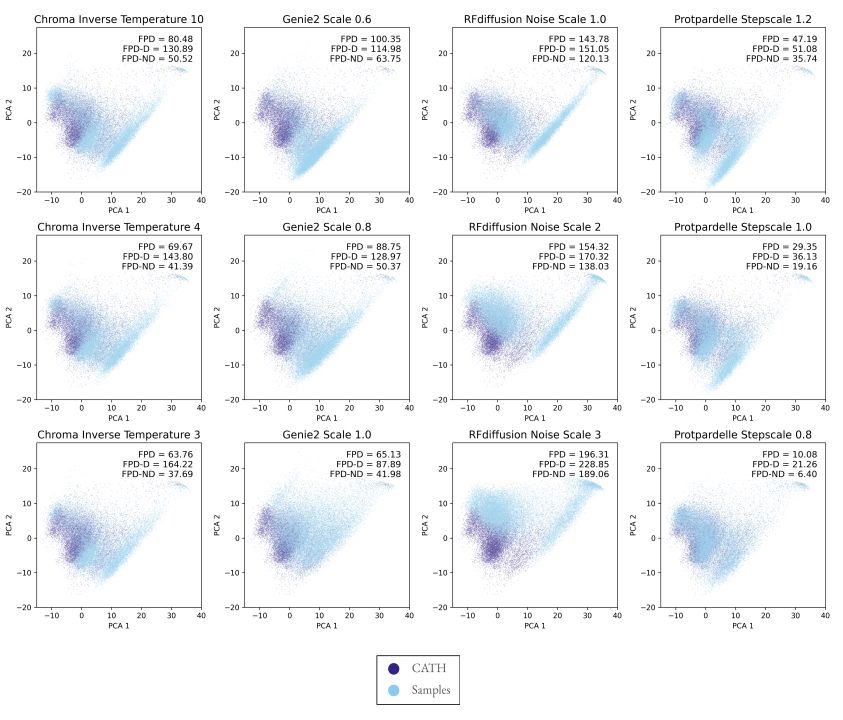
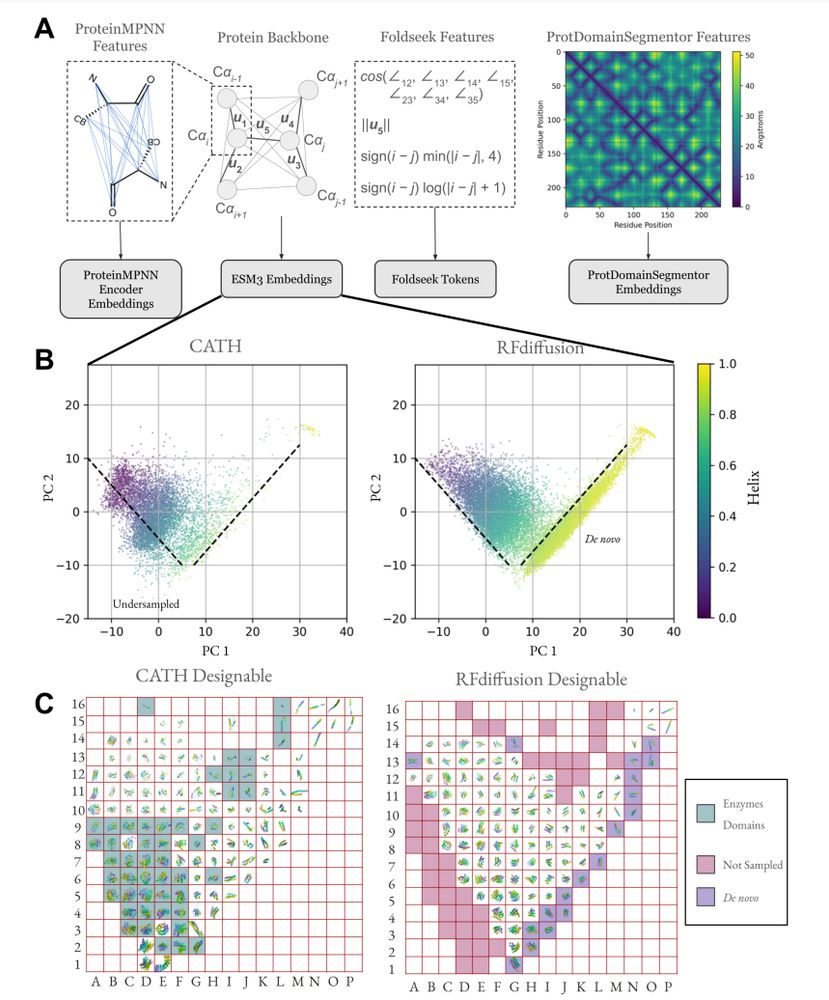
@possuhuanglab.bsky.social
www.biorxiv.org/content/10.1...

Congratulations to @haotiandu.bsky.social and @possuhuanglab.bsky.social on the 2 papers!
TRACeR for MHC-I: go.nature.com/4gcLzn5
TRACeR for MHC-II: go.nature.com/4gj5OQk

Congratulations to @haotiandu.bsky.social and @possuhuanglab.bsky.social on the 2 papers!
TRACeR for MHC-I: go.nature.com/4gcLzn5
TRACeR for MHC-II: go.nature.com/4gj5OQk

@possuhuanglab.bsky.social @haotiandu.bsky.social
@possuhuanglab.bsky.social @haotiandu.bsky.social
TRACeR for MHC-I: go.nature.com/4gcLzn5
TRACeR for MHC-II: go.nature.com/4gj5OQk

TRACeR for MHC-I: go.nature.com/4gcLzn5
TRACeR for MHC-II: go.nature.com/4gj5OQk

It powers CAR-T and BiTE therapies, showing potential to transform cancer and infectious disease treatment.
www.nature.com/articles/s41...
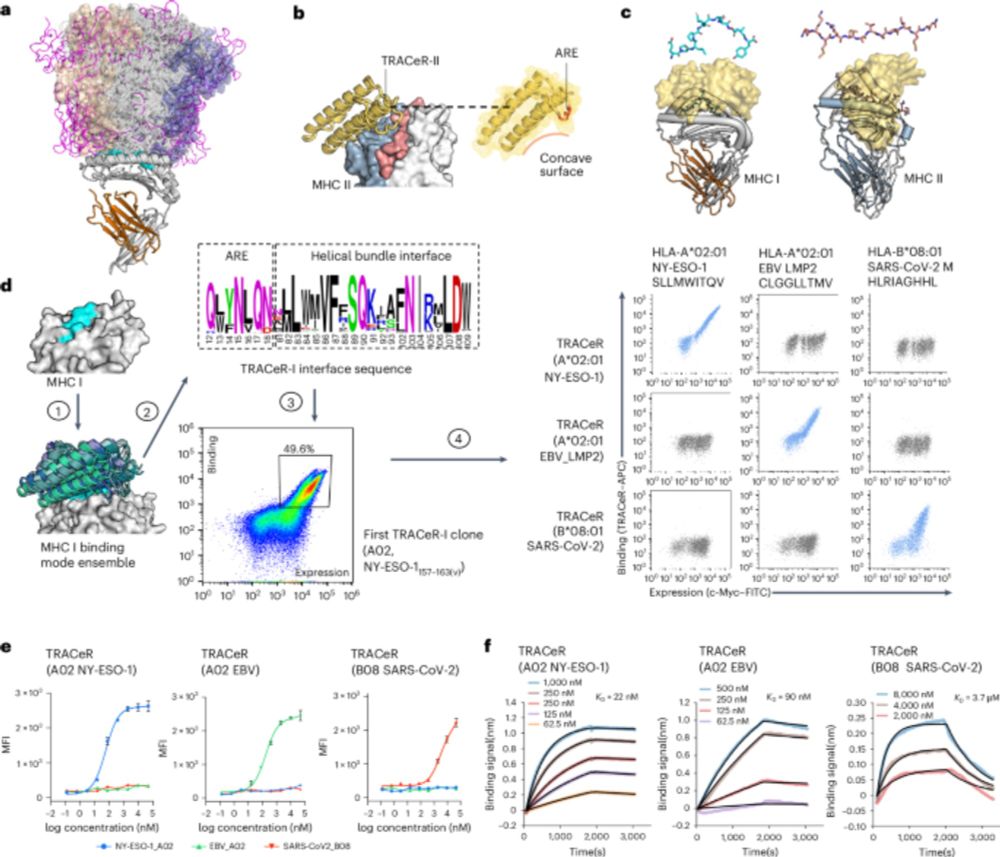
It powers CAR-T and BiTE therapies, showing potential to transform cancer and infectious disease treatment.
www.nature.com/articles/s41...
TRACeR for MHC-I: go.nature.com/4gcLzn5
TRACeR for MHC-II: go.nature.com/4gj5OQk

TRACeR for MHC-I: go.nature.com/4gcLzn5
TRACeR for MHC-II: go.nature.com/4gj5OQk

