Possu Huang Lab
@possuhuanglab.bsky.social
Our lab uses experimental and computational methods to design de novo proteins | @Stanford
Read our preprint here: www.biorxiv.org/content/10.1... (8/8)

SLAE: Strictly Local All-atom Environment for Protein Representation
Building physically grounded protein representations is central to computational biology, yet most existing approaches rely on sequence-pretrained language models or backbone-only graphs that overlook...
www.biorxiv.org
October 9, 2025 at 5:36 PM
Read our preprint here: www.biorxiv.org/content/10.1... (8/8)
Work done by Yilin Chen, @tianyu.bsky.social , Cizhang Zhao and @hkws.bsky.social . Thank you all! (7/8)
October 9, 2025 at 5:36 PM
Work done by Yilin Chen, @tianyu.bsky.social , Cizhang Zhao and @hkws.bsky.social . Thank you all! (7/8)
SLAE projects all-atom structures onto a smooth manifold! Unguided linear interpolation between conformations in SLAE latent space decodes to coherent intermediates structures. (6/8)
October 9, 2025 at 5:36 PM
SLAE projects all-atom structures onto a smooth manifold! Unguided linear interpolation between conformations in SLAE latent space decodes to coherent intermediates structures. (6/8)
SLAE extends our generative coverage assessment SHAPES to all-atom, per-residue-type granularity. Now we can compare de novo all-atom protein design models and spot residue-level environment biases. (5/8)

October 9, 2025 at 5:36 PM
SLAE extends our generative coverage assessment SHAPES to all-atom, per-residue-type granularity. Now we can compare de novo all-atom protein design models and spot residue-level environment biases. (5/8)
Rich in atomic-environment signal, SLAE features outperform PLMs and task-specific models across diverse, challenging downstream tasks, including binding affinity, thermostability and chemical shift prediction. All-atom structure pretraining is all you need! (4/8)

October 9, 2025 at 5:36 PM
Rich in atomic-environment signal, SLAE features outperform PLMs and task-specific models across diverse, challenging downstream tasks, including binding affinity, thermostability and chemical shift prediction. All-atom structure pretraining is all you need! (4/8)
The SLAE latent landscape is organized in meaningful ways beyond amino acid identity. It separates residue embeddings along features including solvent accessibility, secondary structure and structural nativeness. (3/8)

October 9, 2025 at 5:36 PM
The SLAE latent landscape is organized in meaningful ways beyond amino acid identity. It separates residue embeddings along features including solvent accessibility, secondary structure and structural nativeness. (3/8)
We design a deliberately hard two-part task to learn compact, expressive features: a local graph encoder projects each residue’s atomic interactions into a feature vector, while a global decoder learns to compose these local environment tokens into coherent macromolecules. (2/8)
October 9, 2025 at 5:36 PM
We design a deliberately hard two-part task to learn compact, expressive features: a local graph encoder projects each residue’s atomic interactions into a feature vector, while a global decoder learns to compose these local environment tokens into coherent macromolecules. (2/8)
Code will be released soon on our GitHub: github.com/ProteinDesig...
Preprint: www.biorxiv.org/content/10.1...
Have fun sampling and training!
Preprint: www.biorxiv.org/content/10.1...
Have fun sampling and training!

Conditional Protein Structure Generation with Protpardelle-1c
We present Protpardelle-1c, a collection of protein structure generative models with robust motif scaffolding and support for multi-chain complex generation under hotspot-conditioning. Enabling sidech...
www.biorxiv.org
August 19, 2025 at 5:16 PM
Code will be released soon on our GitHub: github.com/ProteinDesig...
Preprint: www.biorxiv.org/content/10.1...
Have fun sampling and training!
Preprint: www.biorxiv.org/content/10.1...
Have fun sampling and training!
Our new set of all-atom models can sample plausible sidechains without stage-2 sampling. Sequence-dependent partial diffusion behavior occurs when we mask the dummy atoms.

August 19, 2025 at 5:16 PM
Our new set of all-atom models can sample plausible sidechains without stage-2 sampling. Sequence-dependent partial diffusion behavior occurs when we mask the dummy atoms.
We achieve competitive results on MotifBench and the RFdiffusion/La-Proteina motif scaffolding benchmarks with both backbone-only and all-atom models, proposing scaffolds to previously unsolved problems.

August 19, 2025 at 5:16 PM
We achieve competitive results on MotifBench and the RFdiffusion/La-Proteina motif scaffolding benchmarks with both backbone-only and all-atom models, proposing scaffolds to previously unsolved problems.
July 29, 2025 at 7:04 PM
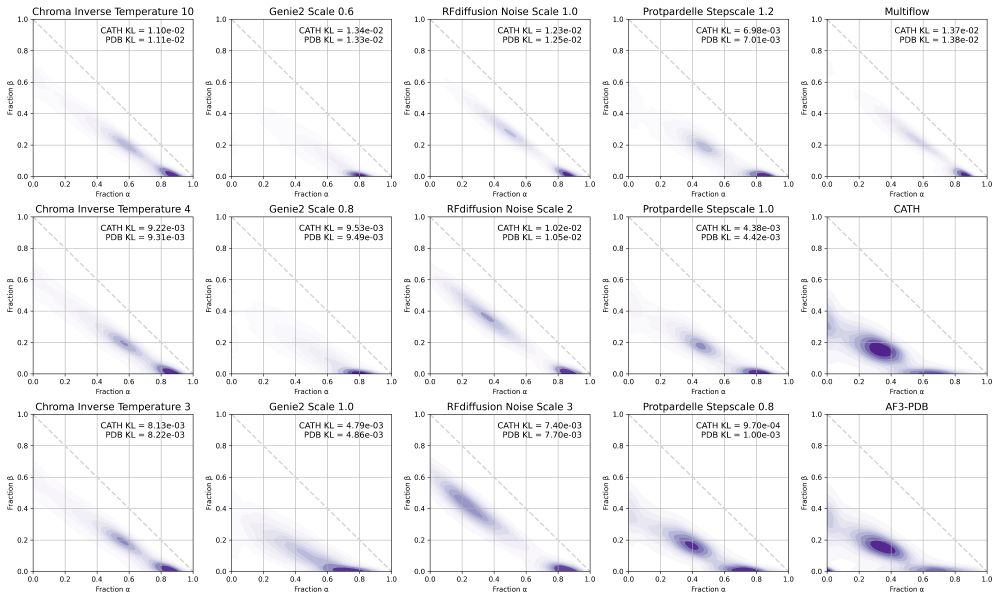
We include some additional analysis in the supplement, including secondary structure distributions.
July 29, 2025 at 7:04 PM
We include some additional analysis in the supplement, including secondary structure distributions.
Preprint: www.biorxiv.org/content/10.1...
Code: github.com/ProteinDesig...
Dataset: zenodo.org/records/1458...
Code: github.com/ProteinDesig...
Dataset: zenodo.org/records/1458...
January 15, 2025 at 6:48 PM
Preprint: www.biorxiv.org/content/10.1...
Code: github.com/ProteinDesig...
Dataset: zenodo.org/records/1458...
Code: github.com/ProteinDesig...
Dataset: zenodo.org/records/1458...
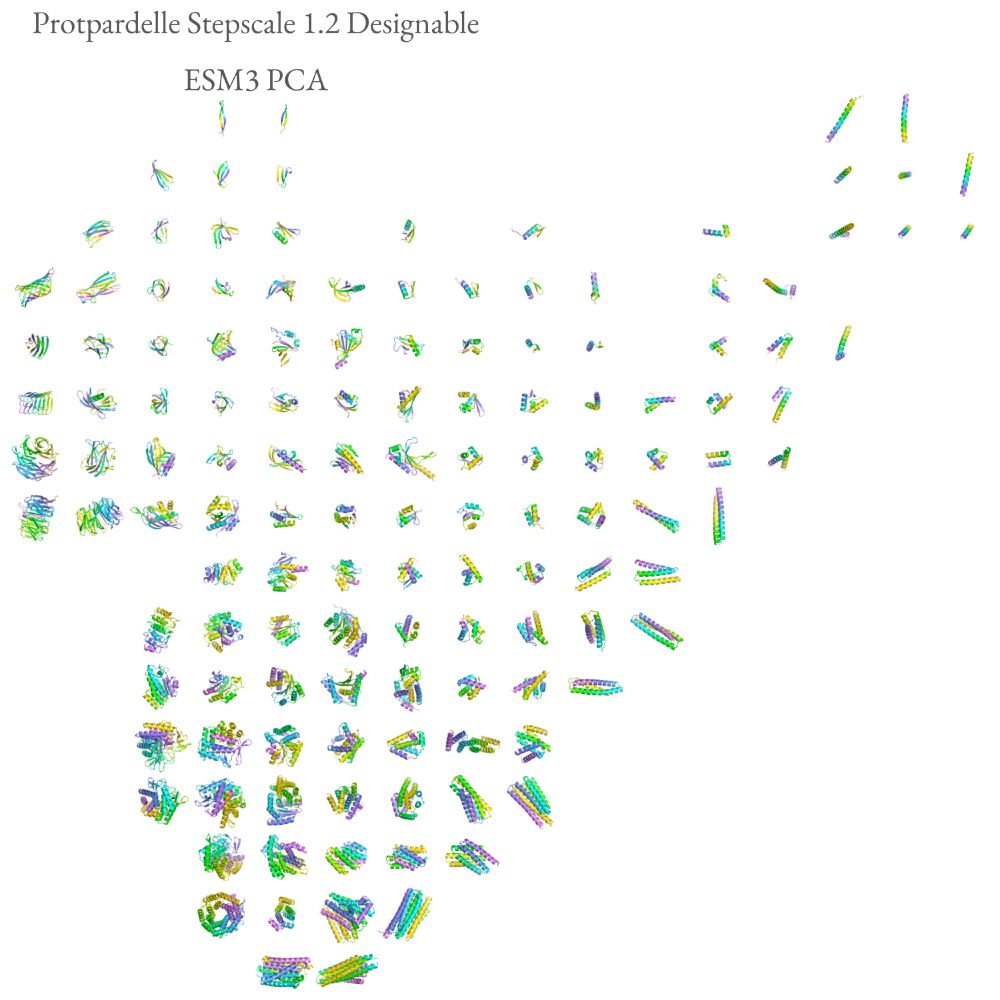
Our supplement has many additional figures of the rasterized protein structure space, stratified by designable and not designable and spatially organized by ESM3 and ProtDomainSegmentor embeddings.
January 15, 2025 at 6:48 PM
Our supplement has many additional figures of the rasterized protein structure space, stratified by designable and not designable and spatially organized by ESM3 and ProtDomainSegmentor embeddings.
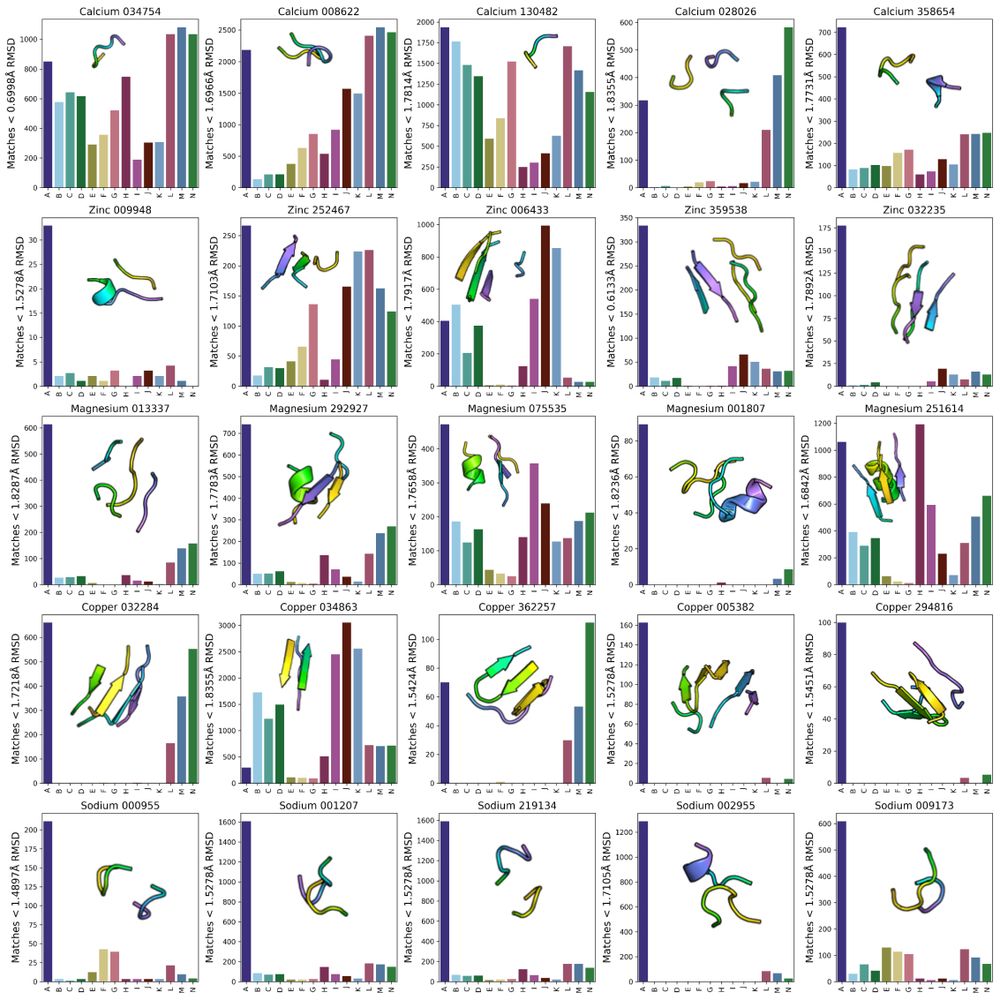
One consequence of unbiased sampling of protein structure space is a higher likelihood of finding TERtiary Motifs (TERMs) which involve complex loops, with implications for functional protein design (see Figure 5 legend for group labels).
January 15, 2025 at 6:48 PM
One consequence of unbiased sampling of protein structure space is a higher likelihood of finding TERtiary Motifs (TERMs) which involve complex loops, with implications for functional protein design (see Figure 5 legend for group labels).
Inspired by the FPD metric in EvoDiff for protein sequence distributions, we compute Fréchet distance using protein structure embeddings, also subsetted to designable and non-designable samples (FPD-D and FPD-ND).
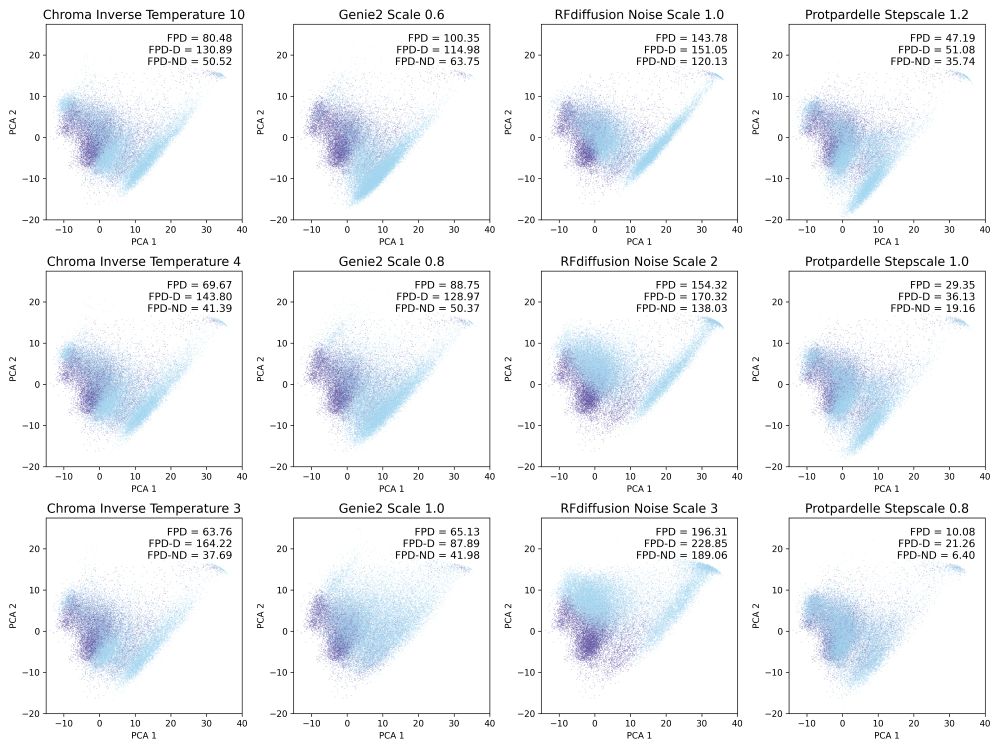
January 15, 2025 at 6:48 PM
Inspired by the FPD metric in EvoDiff for protein sequence distributions, we compute Fréchet distance using protein structure embeddings, also subsetted to designable and non-designable samples (FPD-D and FPD-ND).


