Possu Huang Lab
@possuhuanglab.bsky.social
Our lab uses experimental and computational methods to design de novo proteins | @Stanford
SLAE projects all-atom structures onto a smooth manifold! Unguided linear interpolation between conformations in SLAE latent space decodes to coherent intermediates structures. (6/8)
October 9, 2025 at 5:36 PM
SLAE projects all-atom structures onto a smooth manifold! Unguided linear interpolation between conformations in SLAE latent space decodes to coherent intermediates structures. (6/8)
SLAE extends our generative coverage assessment SHAPES to all-atom, per-residue-type granularity. Now we can compare de novo all-atom protein design models and spot residue-level environment biases. (5/8)

October 9, 2025 at 5:36 PM
SLAE extends our generative coverage assessment SHAPES to all-atom, per-residue-type granularity. Now we can compare de novo all-atom protein design models and spot residue-level environment biases. (5/8)
Rich in atomic-environment signal, SLAE features outperform PLMs and task-specific models across diverse, challenging downstream tasks, including binding affinity, thermostability and chemical shift prediction. All-atom structure pretraining is all you need! (4/8)

October 9, 2025 at 5:36 PM
Rich in atomic-environment signal, SLAE features outperform PLMs and task-specific models across diverse, challenging downstream tasks, including binding affinity, thermostability and chemical shift prediction. All-atom structure pretraining is all you need! (4/8)
The SLAE latent landscape is organized in meaningful ways beyond amino acid identity. It separates residue embeddings along features including solvent accessibility, secondary structure and structural nativeness. (3/8)

October 9, 2025 at 5:36 PM
The SLAE latent landscape is organized in meaningful ways beyond amino acid identity. It separates residue embeddings along features including solvent accessibility, secondary structure and structural nativeness. (3/8)
Introducing SLAE, our new framework to represent all-atom protein structures with residue local chemical environment tokens!
SLAE reasons over atomic interactions to recover structures and residue pairwise energetics, yielding a generalizable, physics-informed latent space. (1/8)
SLAE reasons over atomic interactions to recover structures and residue pairwise energetics, yielding a generalizable, physics-informed latent space. (1/8)

October 9, 2025 at 5:36 PM
Introducing SLAE, our new framework to represent all-atom protein structures with residue local chemical environment tokens!
SLAE reasons over atomic interactions to recover structures and residue pairwise energetics, yielding a generalizable, physics-informed latent space. (1/8)
SLAE reasons over atomic interactions to recover structures and residue pairwise energetics, yielding a generalizable, physics-informed latent space. (1/8)
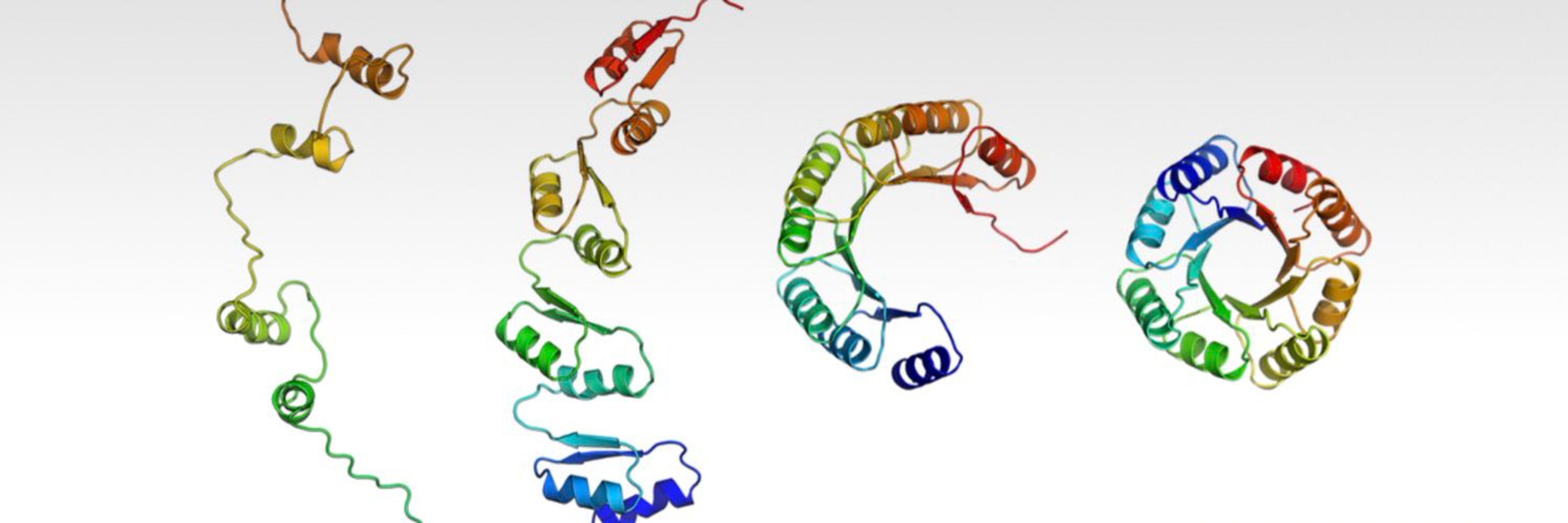
Our new set of all-atom models can sample plausible sidechains without stage-2 sampling. Sequence-dependent partial diffusion behavior occurs when we mask the dummy atoms.

August 19, 2025 at 5:16 PM
Our new set of all-atom models can sample plausible sidechains without stage-2 sampling. Sequence-dependent partial diffusion behavior occurs when we mask the dummy atoms.
We achieve competitive results on MotifBench and the RFdiffusion/La-Proteina motif scaffolding benchmarks with both backbone-only and all-atom models, proposing scaffolds to previously unsolved problems.

August 19, 2025 at 5:16 PM
We achieve competitive results on MotifBench and the RFdiffusion/La-Proteina motif scaffolding benchmarks with both backbone-only and all-atom models, proposing scaffolds to previously unsolved problems.
We have a new collection of protein structure generative models which we call Protpardelle-1c. It builds on the original Protpardelle and is tailored for conditional generation: motif scaffolding and binder generation.

August 19, 2025 at 5:16 PM
We have a new collection of protein structure generative models which we call Protpardelle-1c. It builds on the original Protpardelle and is tailored for conditional generation: motif scaffolding and binder generation.
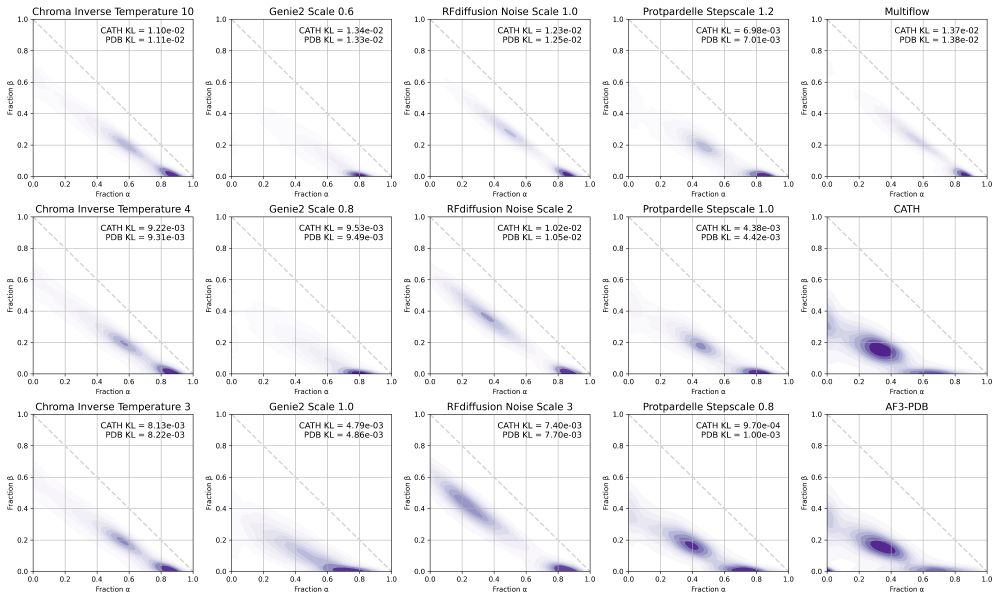
We include some additional analysis in the supplement, including secondary structure distributions.
July 29, 2025 at 7:04 PM
We include some additional analysis in the supplement, including secondary structure distributions.
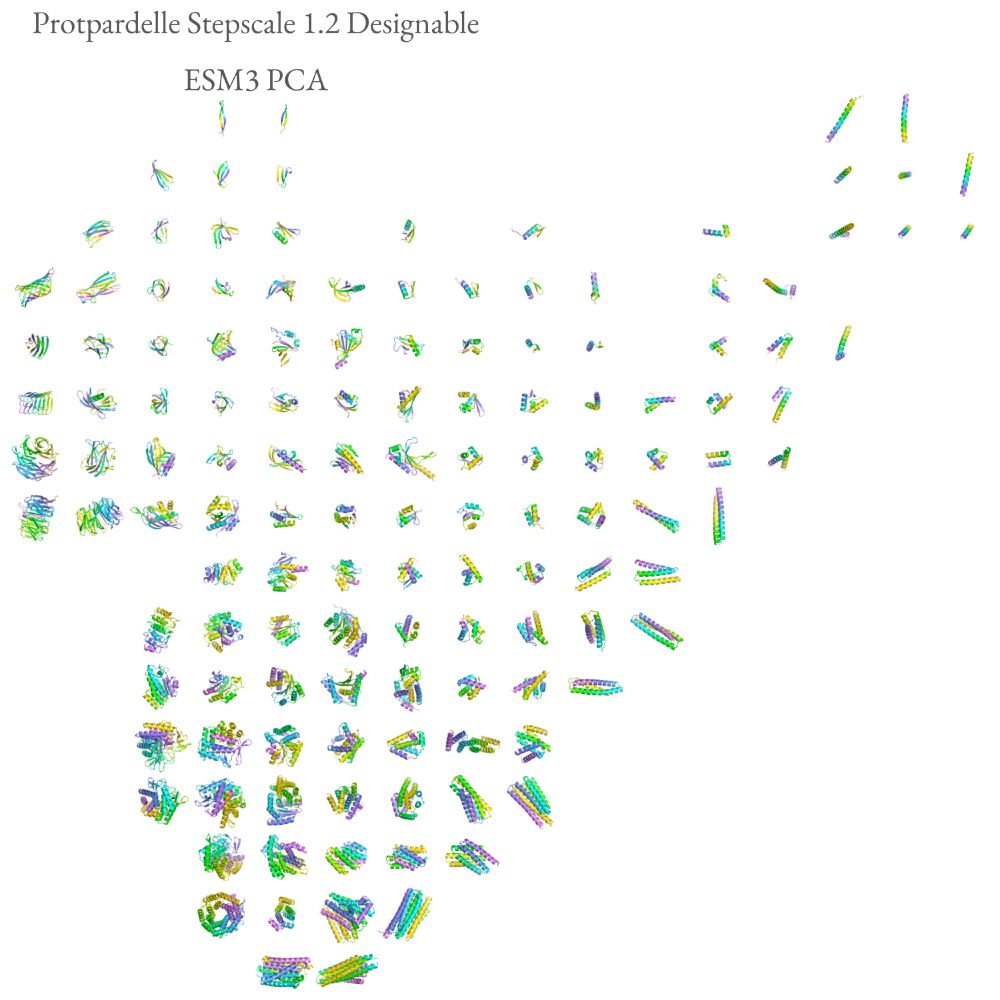
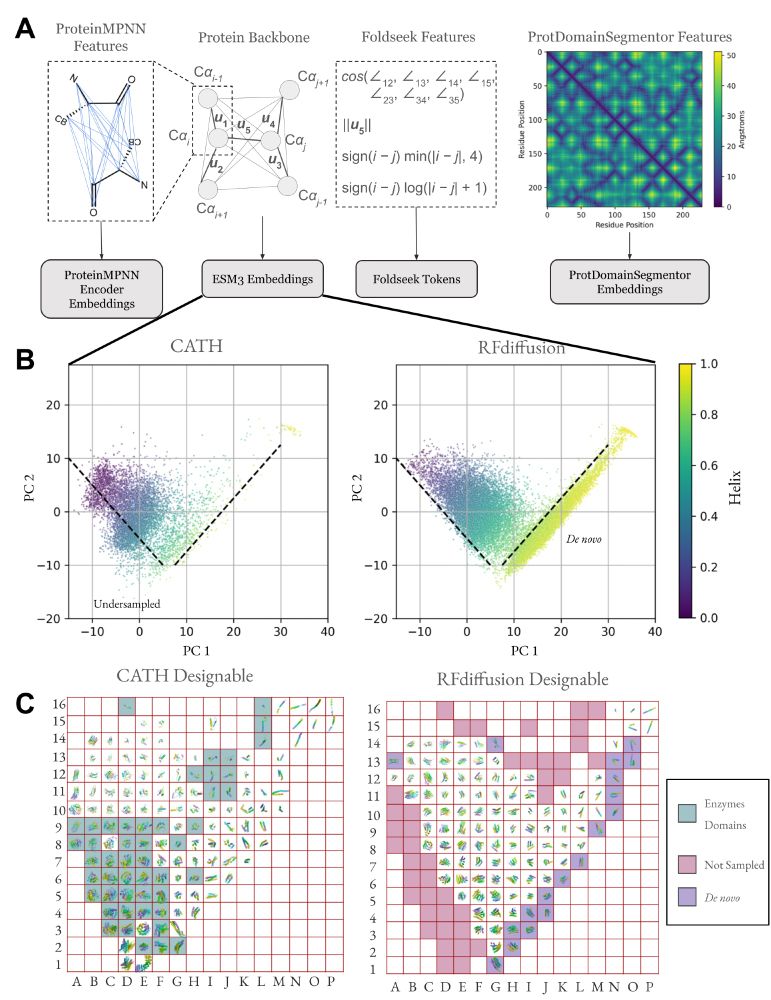
Our supplement has many additional figures of the rasterized protein structure space, stratified by designable and not designable and spatially organized by ESM3 and ProtDomainSegmentor embeddings.
January 15, 2025 at 6:48 PM
Our supplement has many additional figures of the rasterized protein structure space, stratified by designable and not designable and spatially organized by ESM3 and ProtDomainSegmentor embeddings.
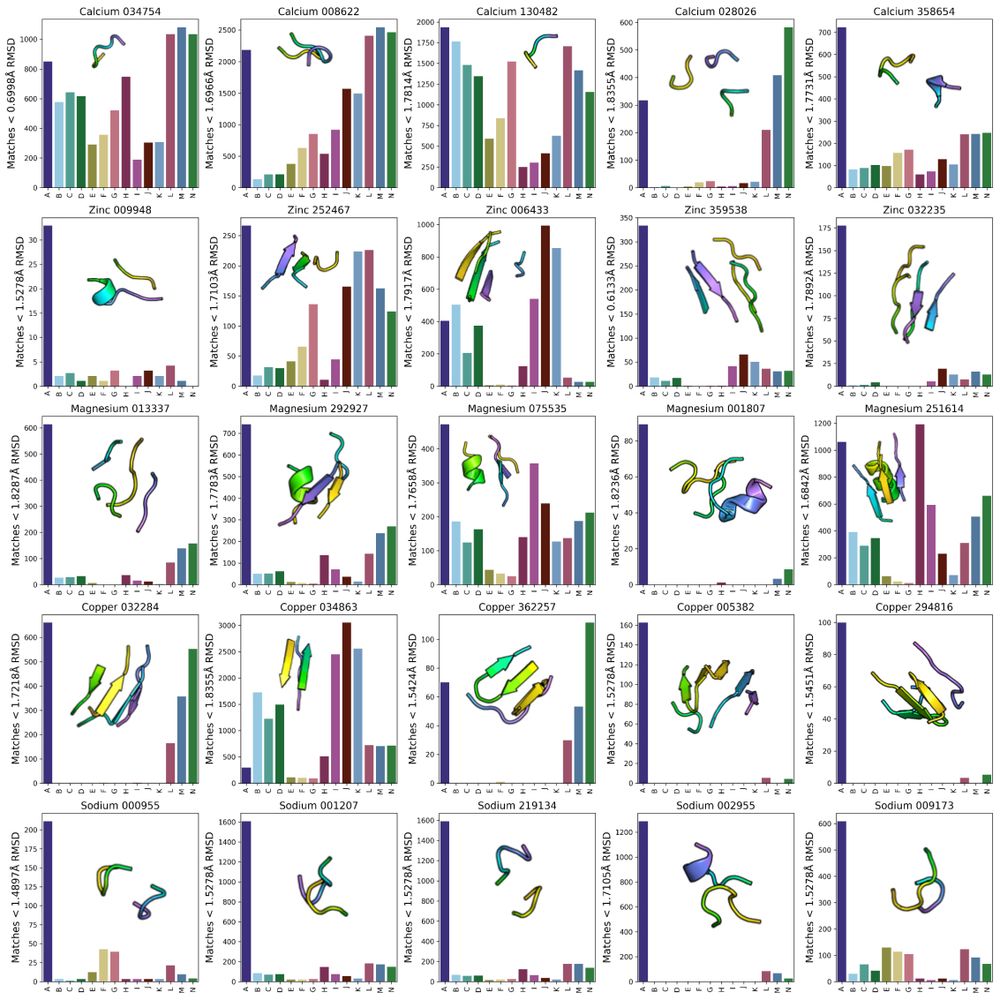
One consequence of unbiased sampling of protein structure space is a higher likelihood of finding TERtiary Motifs (TERMs) which involve complex loops, with implications for functional protein design (see Figure 5 legend for group labels).
January 15, 2025 at 6:48 PM
One consequence of unbiased sampling of protein structure space is a higher likelihood of finding TERtiary Motifs (TERMs) which involve complex loops, with implications for functional protein design (see Figure 5 legend for group labels).
Inspired by the FPD metric in EvoDiff for protein sequence distributions, we compute Fréchet distance using protein structure embeddings, also subsetted to designable and non-designable samples (FPD-D and FPD-ND).
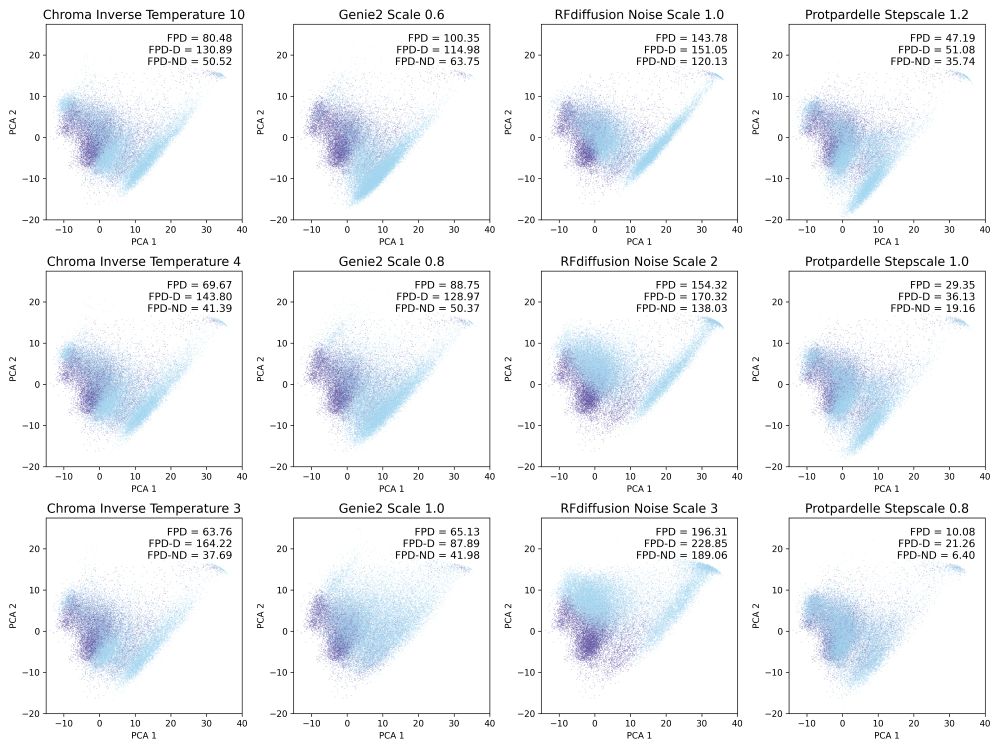
January 15, 2025 at 6:48 PM
Inspired by the FPD metric in EvoDiff for protein sequence distributions, we compute Fréchet distance using protein structure embeddings, also subsetted to designable and non-designable samples (FPD-D and FPD-ND).
New preprint from our group! We propose SHAPES, a set of metrics to quantify the distributional coverage of generative models of protein structures with embeddings at different structural hierarchies and quantify undersampling / extrapolation behaviors.
January 15, 2025 at 6:48 PM
New preprint from our group! We propose SHAPES, a set of metrics to quantify the distributional coverage of generative models of protein structures with embeddings at different structural hierarchies and quantify undersampling / extrapolation behaviors.
7/ Our platform offers a convenient strategy for creating MHC binders: from a master library, peptide-specific MHC binders can be isolated simply by a few rounds of yeast surface display enrichment.

December 17, 2024 at 12:56 AM
7/ Our platform offers a convenient strategy for creating MHC binders: from a master library, peptide-specific MHC binders can be isolated simply by a few rounds of yeast surface display enrichment.
6/ We show proof-of-concept targeted killing of tumor cells using TRACeR-I.
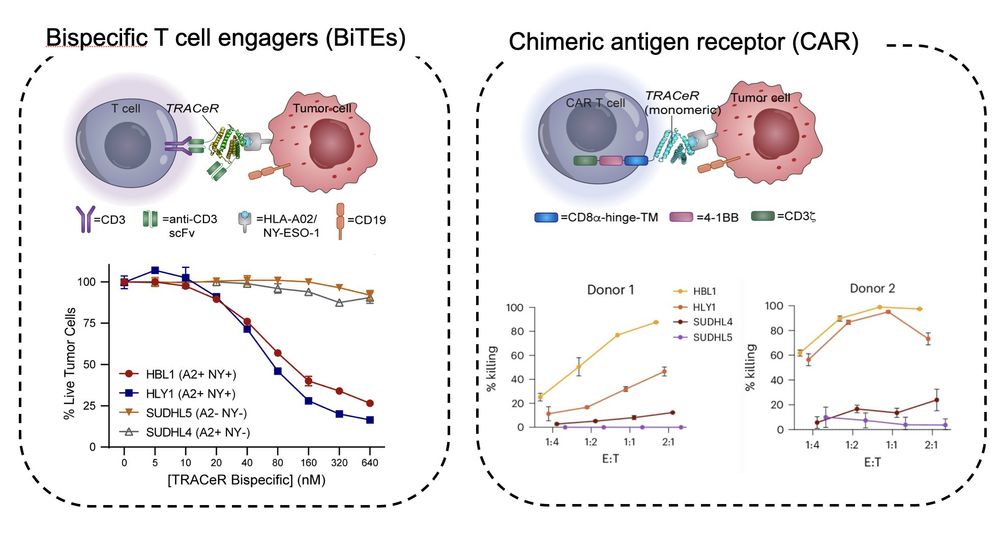
December 17, 2024 at 12:56 AM
6/ We show proof-of-concept targeted killing of tumor cells using TRACeR-I.
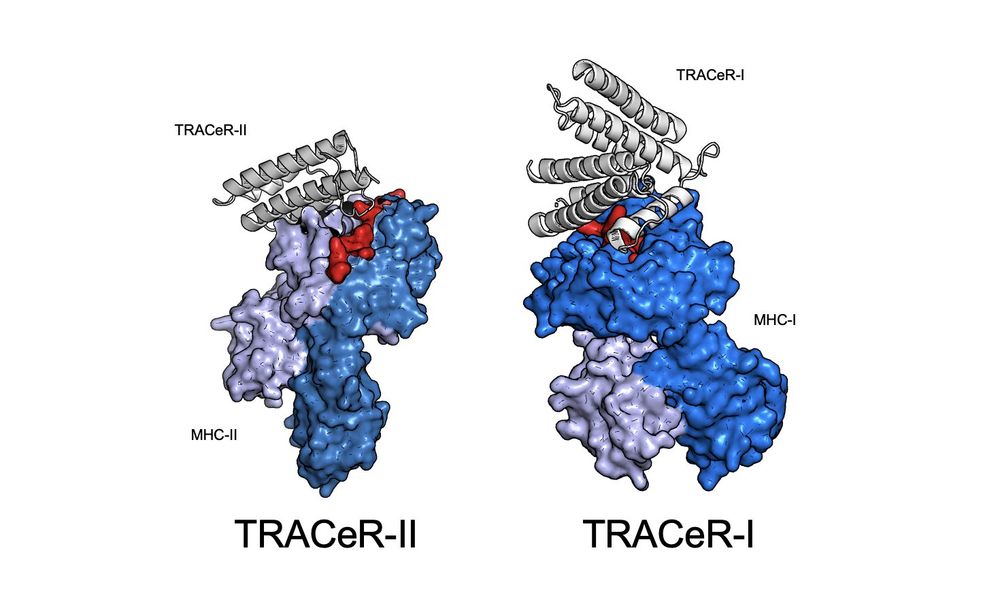
5/ The mechanism of TRACeR-1 can achieve discrimination of single point mutants to peptides.
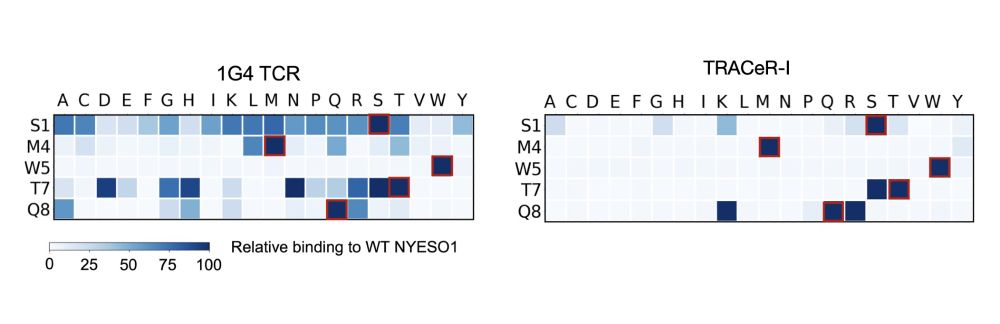
December 17, 2024 at 12:56 AM
5/ The mechanism of TRACeR-1 can achieve discrimination of single point mutants to peptides.
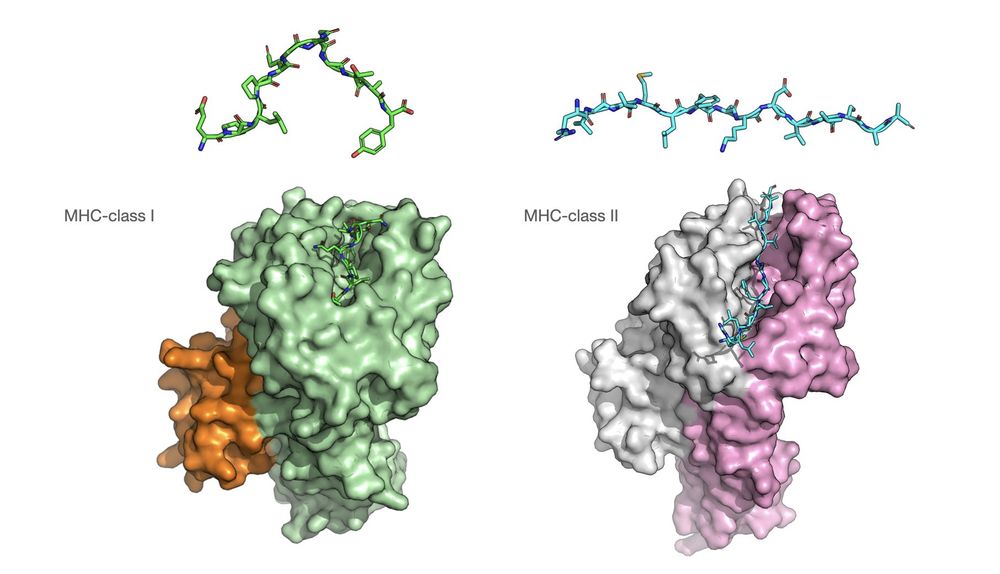
4/ But MHC-I peptides have bulged conformations. For TRACeR-I, we use a set of spatially-defined amino acid residues to “box-in” on the peptide and achieve recognition.

December 17, 2024 at 12:56 AM
4/ But MHC-I peptides have bulged conformations. For TRACeR-I, we use a set of spatially-defined amino acid residues to “box-in” on the peptide and achieve recognition.
3/ Peptides in MHC-II are in an extended conformation. TRACeR-II uses a surface loop to reach into MHC-II’s peptide binding groove to recognize the peptide. We use a single loop to convey specificity to the peptide in the binding groove.

December 17, 2024 at 12:56 AM
3/ Peptides in MHC-II are in an extended conformation. TRACeR-II uses a surface loop to reach into MHC-II’s peptide binding groove to recognize the peptide. We use a single loop to convey specificity to the peptide in the binding groove.
2/ MHC antigens are central to the immune system and can be leveraged for disease-specific targeting. These peptides are derived from intra-cellular proteins and ingested foreign antigens for presentation by class-I and class-II MHCs, respectively.
December 17, 2024 at 12:56 AM
2/ MHC antigens are central to the immune system and can be leveraged for disease-specific targeting. These peptides are derived from intra-cellular proteins and ingested foreign antigens for presentation by class-I and class-II MHCs, respectively.
1/ In two back-to-back papers, we present our de novo TRACeR platform for targeting MHC-I and MHC-II antigens
TRACeR for MHC-I: go.nature.com/4gcLzn5
TRACeR for MHC-II: go.nature.com/4gj5OQk
TRACeR for MHC-I: go.nature.com/4gcLzn5
TRACeR for MHC-II: go.nature.com/4gj5OQk
December 17, 2024 at 12:56 AM
1/ In two back-to-back papers, we present our de novo TRACeR platform for targeting MHC-I and MHC-II antigens
TRACeR for MHC-I: go.nature.com/4gcLzn5
TRACeR for MHC-II: go.nature.com/4gj5OQk
TRACeR for MHC-I: go.nature.com/4gcLzn5
TRACeR for MHC-II: go.nature.com/4gj5OQk


