
https://scholar.google.co.jp/citations?user=Q34_N9wAAAAJ&hl=ja#
www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...
www.biorxiv.org/content/10.6...
www.biorxiv.org/content/10.6...
👉 buff.ly/ApWkCtg

👉 buff.ly/ApWkCtg
@buzzbaum.bsky.social et al define mechanisms underlying switch-like degradation of archaeal ESCRT-III CvdB in Sulfolobus
Thanks to @reviewcommons.org for another #RefereedPreprint
link.springer.com/article/10.1...

@buzzbaum.bsky.social et al define mechanisms underlying switch-like degradation of archaeal ESCRT-III CvdB in Sulfolobus
Thanks to @reviewcommons.org for another #RefereedPreprint
link.springer.com/article/10.1...
www.ls.tum.de/en/ls/public...

www.ls.tum.de/en/ls/public...
www.sciencedirect.com/science/arti...

www.sciencedirect.com/science/arti...
doi.org/10.1247/csf....

doi.org/10.1247/csf....
www.nature.com/articles/s41...

www.nature.com/articles/s41...
www.nature.com/articles/s41...
www.nature.com/articles/s41...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
Many thanks to the organizers — looking forward to inspiring talks and discussions in Cambridge!
meetings.embo.org/event/26-arc...
Many thanks to the organizers — looking forward to inspiring talks and discussions in Cambridge!
meetings.embo.org/event/26-arc...
Either way, we have good news.
We’re delighted to announce the 2026 EMBO Workshop on Archaea, 6–10 July.
Sign up: meetings.embo.org/event/26-arc...
We look forward to seeing you in Cambridge, UK.
Please repost!
Either way, we have good news.
We’re delighted to announce the 2026 EMBO Workshop on Archaea, 6–10 July.
Sign up: meetings.embo.org/event/26-arc...
We look forward to seeing you in Cambridge, UK.
Please repost!
We address a longstanding question about the role of the topoisomerase reverse gyrase in hyperthermophiles. Great collaboration with Baranello and @tobiaswarnecke.bsky.social groups. #Archaeasky #Microsky

We address a longstanding question about the role of the topoisomerase reverse gyrase in hyperthermophiles. Great collaboration with Baranello and @tobiaswarnecke.bsky.social groups. #Archaeasky #Microsky
www.biorxiv.org/content/10.6...

www.biorxiv.org/content/10.6...
Nous recrutons un/e Ingénieur/e d’étude en biologie.
Profil : être titulaire d’un poste permanent au CNRS (IE) et avoir envie d’étudier la chromatine bactérienne 😉
Date limite : jeudi 15 janvier.
Détails et candidature ici :
mobiliteinterne.cnrs.fr/ords/afip/ow...
Nous recrutons un/e Ingénieur/e d’étude en biologie.
Profil : être titulaire d’un poste permanent au CNRS (IE) et avoir envie d’étudier la chromatine bactérienne 😉
Date limite : jeudi 15 janvier.
Détails et candidature ici :
mobiliteinterne.cnrs.fr/ords/afip/ow...
Face-to-face histones are important organizers of archaeal chromatin alongside classical histones.
www.biorxiv.org/content/10.6...

www.findaphd.com/phds/project...
📅 January 7, 2026
Please RP🙏thx!


www.findaphd.com/phds/project...
📅 January 7, 2026
Please RP🙏thx!
Asgard histones form closed and open hypernucleosomes. Closed are conserved across #Archaea, while open resemble eukaryotic H3–H4 octasomes and are Asgard-specific. More here: www.cell.com/molecular-ce...

Asgard histones form closed and open hypernucleosomes. Closed are conserved across #Archaea, while open resemble eukaryotic H3–H4 octasomes and are Asgard-specific. More here: www.cell.com/molecular-ce...
Free access link: rdcu.be/eLtCH
🧵 by @yifanzhou.bsky.social 👇
@mkrupovic.bsky.social @deemteam.bsky.social @anagtz.bsky.social
www.nature.com/articles/s41...

Free access link: rdcu.be/eLtCH
🧵 by @yifanzhou.bsky.social 👇
doi.org/10.1073/pnas...

doi.org/10.1073/pnas...
www.nature.com/articles/s41...
Thanks again to our awesome collaborators @mblokesch.bsky.social and David and co and Mark Szczelkun and @steven-shaw.bsky.social and the DCI Lausanne @fbm-unil.bsky.social
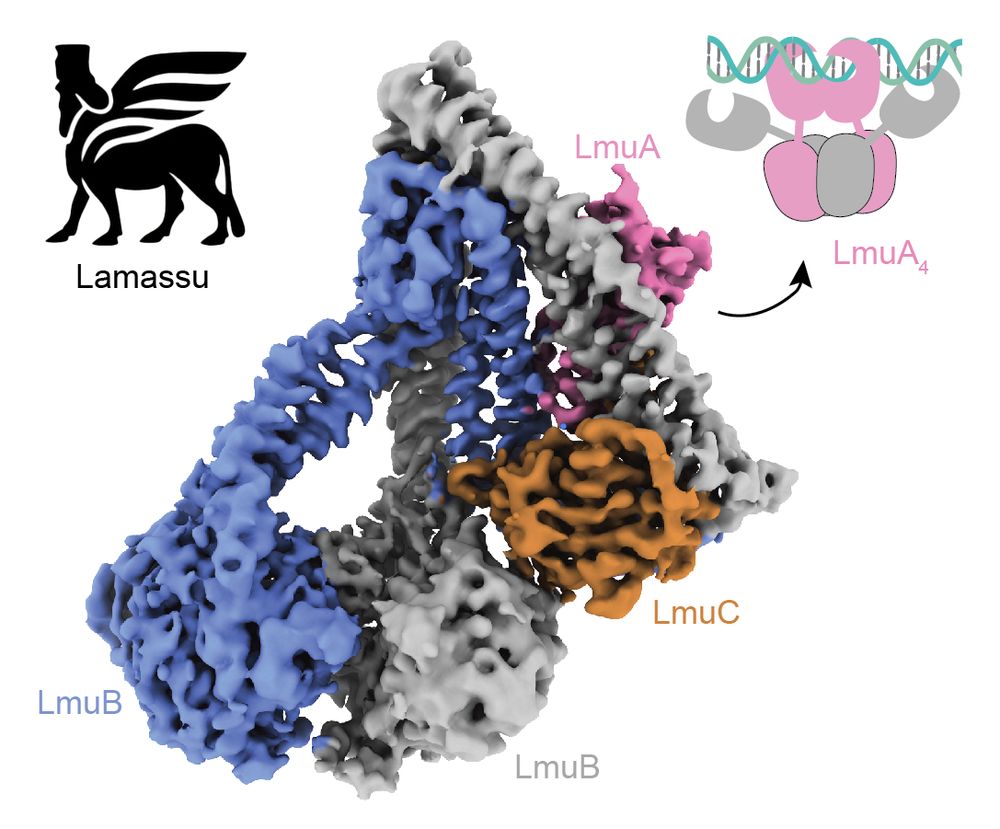
www.nature.com/articles/s41...
Thanks again to our awesome collaborators @mblokesch.bsky.social and David and co and Mark Szczelkun and @steven-shaw.bsky.social and the DCI Lausanne @fbm-unil.bsky.social
A fantastic collaboration with Antoine, with Jovana Kaljevic' initiated the collaboration and drives the project.

A fantastic collaboration with Antoine, with Jovana Kaljevic' initiated the collaboration and drives the project.

