Luca Giorgetti lab @FMI
@lucagiorgetti.bsky.social
We study transcriptional regulation and chromosome folding using an interdisciplinary approach combining wet- and dry-lab methods.
https://giorgettilab.org
@fmiscience.bsky.social
https://giorgettilab.org
@fmiscience.bsky.social
Pinned
Really excited to share our latest work led by @mattiaubertini.bsky.social and @nesslfy.bsky.social: we report that cohesin loop extrusion creates rare but long-lived encounters between genomic sequences which underlie efficient enhancer-promoter communication.
www.biorxiv.org/content/10.1...
A🧵👇
www.biorxiv.org/content/10.1...
A🧵👇
Reposted by Luca Giorgetti lab @FMI
Our collab w. V Goel, @nicholas-aboreden.bsky.social , J Jusuf, G Blobel, L Mirny, @irate-physicist.bsky.social out in @natsmb.nature.com www.nature.com/articles/s41...
Was co-submitted with @allanaschooley.bsky.social @jobdekker.bsky.social whose paper should also come out soon
Brief thread 👇
Was co-submitted with @allanaschooley.bsky.social @jobdekker.bsky.social whose paper should also come out soon
Brief thread 👇
October 20, 2025 at 6:36 PM
Our collab w. V Goel, @nicholas-aboreden.bsky.social , J Jusuf, G Blobel, L Mirny, @irate-physicist.bsky.social out in @natsmb.nature.com www.nature.com/articles/s41...
Was co-submitted with @allanaschooley.bsky.social @jobdekker.bsky.social whose paper should also come out soon
Brief thread 👇
Was co-submitted with @allanaschooley.bsky.social @jobdekker.bsky.social whose paper should also come out soon
Brief thread 👇
Reposted by Luca Giorgetti lab @FMI
What is a promoter? And how does it work?
We very happy to share our latest work trying to understand enhancer-promoter compatibility.
I am very excited about the results of @blanka-majchrzycka.bsky.social, which changed the way I think about promoters
www.biorxiv.org/content/10.1...
We very happy to share our latest work trying to understand enhancer-promoter compatibility.
I am very excited about the results of @blanka-majchrzycka.bsky.social, which changed the way I think about promoters
www.biorxiv.org/content/10.1...

Enhancer-promoter compatibility is mediated by the promoter-proximal region
Gene promoters induce transcription in response to distal enhancers. How enhancers specifically activate their target promoter while bypassing other promoters remains unclear. Here, we find that the p...
www.biorxiv.org
October 16, 2025 at 3:06 PM
What is a promoter? And how does it work?
We very happy to share our latest work trying to understand enhancer-promoter compatibility.
I am very excited about the results of @blanka-majchrzycka.bsky.social, which changed the way I think about promoters
www.biorxiv.org/content/10.1...
We very happy to share our latest work trying to understand enhancer-promoter compatibility.
I am very excited about the results of @blanka-majchrzycka.bsky.social, which changed the way I think about promoters
www.biorxiv.org/content/10.1...
Reposted by Luca Giorgetti lab @FMI
The TArgeted Cohesin Loader (TACL) paper was just published. Happy that we were able to contribute to this really exciting project!
If you want to learn how targeting cohesin to defined loci in the genome affects the local chromatin environment and transcription, look no further!
rdcu.be/eLiT5
If you want to learn how targeting cohesin to defined loci in the genome affects the local chromatin environment and transcription, look no further!
rdcu.be/eLiT5

Characterization of induced cohesin loop extrusion trajectories in living cells
Nature Genetics - This study introduces a system called TArgeted Cohesin Loader (TACL) that recruits cohesin complexes at defined genomic regions and induces loop extrusion events in living cells,...
rdcu.be
October 16, 2025 at 8:17 PM
The TArgeted Cohesin Loader (TACL) paper was just published. Happy that we were able to contribute to this really exciting project!
If you want to learn how targeting cohesin to defined loci in the genome affects the local chromatin environment and transcription, look no further!
rdcu.be/eLiT5
If you want to learn how targeting cohesin to defined loci in the genome affects the local chromatin environment and transcription, look no further!
rdcu.be/eLiT5
Reposted by Luca Giorgetti lab @FMI
We are recuiting two new Associate Professors here in Oxford Biochemistry. Come join us! Reach out to me if you have any questions. Please repost! tinyurl.com/mr3m7bd3

October 17, 2025 at 2:56 PM
We are recuiting two new Associate Professors here in Oxford Biochemistry. Come join us! Reach out to me if you have any questions. Please repost! tinyurl.com/mr3m7bd3
Reposted by Luca Giorgetti lab @FMI
Come join us in Geneva for everything epigenetics and gene regulation. It will be a great meeting! Please repost!
www.keystonesymposia.org/conferences/...
www.keystonesymposia.org/conferences/...

Epigenetics and Gene Regulation in Health and Disease: Linking Basic Mechanisms with Therapeutic Opportunities | Keystone Symposia
Join us at the Keystone Symposia on Epigenetics and Gene Regulation in Health and Disease: Linking Basic Mechanisms with Therapeutic Opportunities, March 2026, in Geneva, with field leaders!
www.keystonesymposia.org
October 17, 2025 at 2:36 PM
Come join us in Geneva for everything epigenetics and gene regulation. It will be a great meeting! Please repost!
www.keystonesymposia.org/conferences/...
www.keystonesymposia.org/conferences/...
Reposted by Luca Giorgetti lab @FMI
⚠️ Paper alert: Using a novel CRISPR screening approach, we mapped the entire regulatory network controlling Xist—key for X-chromosome inactivation.
👉 We discover how sex and development signals are decoded at a single gene locus.
www.nature.com/articles/s41...
👇 Bluetorial
👉 We discover how sex and development signals are decoded at a single gene locus.
www.nature.com/articles/s41...
👇 Bluetorial

Reporter CRISPR screens decipher cis-regulatory and trans-regulatory principles at the Xist locus - Nature Structural & Molecular Biology
Here Schwämmle et al. develop CRISPR reporter screens to map transcription-factor-regulatory element interactions at the Xist locus, revealing a two-step mechanism integrating developmental and X-dosage signals to initiate X-chromosome inactivation.
www.nature.com
October 6, 2025 at 1:02 PM
⚠️ Paper alert: Using a novel CRISPR screening approach, we mapped the entire regulatory network controlling Xist—key for X-chromosome inactivation.
👉 We discover how sex and development signals are decoded at a single gene locus.
www.nature.com/articles/s41...
👇 Bluetorial
👉 We discover how sex and development signals are decoded at a single gene locus.
www.nature.com/articles/s41...
👇 Bluetorial
Reposted by Luca Giorgetti lab @FMI
Join us at the Hubrecht Institute in Utrecht to start your research group! Fantastic scientific environment, strong support, and nice colleagues :) Apply and please share broadly in your network.
Ready to establish your own research group at the Hubrecht Institute for Molecular and Developmental Biology? We’re seeking a tenure-track group leader to develop an innovative research line within our vibrant scientific community. Learn more & apply! https://f.mtr.cool/jncxfqkuds



September 28, 2025 at 9:43 AM
Join us at the Hubrecht Institute in Utrecht to start your research group! Fantastic scientific environment, strong support, and nice colleagues :) Apply and please share broadly in your network.
Reposted by Luca Giorgetti lab @FMI
The latest work from ours and @vram142.bsky.social lab is out! True teamwork to visualize nascent chromatin with strand resolution, using a fully reconstituted system. Very proud of superstar-PhD student Bruna, and @palindromephd.bsky.social. Learning so much from Vijay’s amazing technologies! RT
Some (+)ve news to lighten another heavy weekend: our latest preprint (c/o Mattiroli + Ramani labs) is up!
www.biorxiv.org/content/10.1...
A tour-de-force by 1st authors Bruna Eckhardt & @palindromephd.bsky.social, focusing on chromatin replication. RTs welcome; tweetorial in 3,2...(1/n)
www.biorxiv.org/content/10.1...
A tour-de-force by 1st authors Bruna Eckhardt & @palindromephd.bsky.social, focusing on chromatin replication. RTs welcome; tweetorial in 3,2...(1/n)

The eukaryotic replisome intrinsically generates asymmetric daughter chromatin fibers
DNA replication is molecularly asymmetric, due to distinct mechanisms for lagging and leading strand DNA synthesis. Whether chromatin assembly on newly replicated strands is also asymmetric remains un...
www.biorxiv.org
September 21, 2025 at 7:12 AM
The latest work from ours and @vram142.bsky.social lab is out! True teamwork to visualize nascent chromatin with strand resolution, using a fully reconstituted system. Very proud of superstar-PhD student Bruna, and @palindromephd.bsky.social. Learning so much from Vijay’s amazing technologies! RT
Reposted by Luca Giorgetti lab @FMI
1/ 🧵 I am excited to share a new preprint from our group:
Single-cell Dam&T-seq during mouse corticogenesis reveals how genome–lamina interactions regulate long neuronal genes.
doi.org/10.1101/2025...
#Neurodevelopment #Epigenetics @hubrechtinstitute.bsky.social @oncodeinstitute.bsky.social
Single-cell Dam&T-seq during mouse corticogenesis reveals how genome–lamina interactions regulate long neuronal genes.
doi.org/10.1101/2025...
#Neurodevelopment #Epigenetics @hubrechtinstitute.bsky.social @oncodeinstitute.bsky.social

MeCP2 binding and genome–lamina reorganization precede long gene activation during mouse corticogenesis
During corticogenesis, neural gene expression is tightly coordinated by chromatin and epigenetic changes, whose misregulation can lead to neurodevelopmental disorders[1][1]–[4][2]. The role of spatial...
doi.org
September 17, 2025 at 8:41 AM
1/ 🧵 I am excited to share a new preprint from our group:
Single-cell Dam&T-seq during mouse corticogenesis reveals how genome–lamina interactions regulate long neuronal genes.
doi.org/10.1101/2025...
#Neurodevelopment #Epigenetics @hubrechtinstitute.bsky.social @oncodeinstitute.bsky.social
Single-cell Dam&T-seq during mouse corticogenesis reveals how genome–lamina interactions regulate long neuronal genes.
doi.org/10.1101/2025...
#Neurodevelopment #Epigenetics @hubrechtinstitute.bsky.social @oncodeinstitute.bsky.social
Really excited to share our latest work led by @mattiaubertini.bsky.social and @nesslfy.bsky.social: we report that cohesin loop extrusion creates rare but long-lived encounters between genomic sequences which underlie efficient enhancer-promoter communication.
www.biorxiv.org/content/10.1...
A🧵👇
www.biorxiv.org/content/10.1...
A🧵👇
September 24, 2025 at 9:45 PM
Really excited to share our latest work led by @mattiaubertini.bsky.social and @nesslfy.bsky.social: we report that cohesin loop extrusion creates rare but long-lived encounters between genomic sequences which underlie efficient enhancer-promoter communication.
www.biorxiv.org/content/10.1...
A🧵👇
www.biorxiv.org/content/10.1...
A🧵👇
Reposted by Luca Giorgetti lab @FMI
New work from my lab with the Brown lab on how genome folding changes during motor neuron maturation, and how this process goes awry in ALS motor neurons. Led by Dr. Ozgun Uyan.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Dynamic changes in chromosome and nuclear architecture during maturation of normal and ALS C9orf72 motor neurons
We have investigated changes in chromosome conformation, nuclear organization, and transcription during differentiation and maturation of control and mutant motor neurons harboring hexanucleotide expa...
www.biorxiv.org
September 23, 2025 at 9:13 PM
New work from my lab with the Brown lab on how genome folding changes during motor neuron maturation, and how this process goes awry in ALS motor neurons. Led by Dr. Ozgun Uyan.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Luca Giorgetti lab @FMI
Another FMI Annual Meeting is in the books! After a night of follies and plenty of dancing, today brought the announcement of our internal prize winners, more inspiring talks, and a vibrant poster session. Already looking forward to next year! 🙌




September 17, 2025 at 11:57 AM
Another FMI Annual Meeting is in the books! After a night of follies and plenty of dancing, today brought the announcement of our internal prize winners, more inspiring talks, and a vibrant poster session. Already looking forward to next year! 🙌
Reposted by Luca Giorgetti lab @FMI
📣The Turco group is seeking a computational biologist to analyze large omics datasets. Work on cutting-edge 3D organoid models of the human placenta and be embedded in our Computational Biology Platform (part of @sib.swiss). Apply here: www.fmi.ch/education-ca...

September 12, 2025 at 7:02 AM
📣The Turco group is seeking a computational biologist to analyze large omics datasets. Work on cutting-edge 3D organoid models of the human placenta and be embedded in our Computational Biology Platform (part of @sib.swiss). Apply here: www.fmi.ch/education-ca...
Reposted by Luca Giorgetti lab @FMI
I am excited to announce that we are looking for a Lab Manager @fmiscience.bsky.social in Basel. If you want to be part of a growing team investigating development and tissue formation, and are enthusiastic about helping set up a new lab, please check out the role:
www.fmi.ch/education-ca...
www.fmi.ch/education-ca...
September 14, 2025 at 11:46 AM
I am excited to announce that we are looking for a Lab Manager @fmiscience.bsky.social in Basel. If you want to be part of a growing team investigating development and tissue formation, and are enthusiastic about helping set up a new lab, please check out the role:
www.fmi.ch/education-ca...
www.fmi.ch/education-ca...
Become our colleague at @fmiscience.bsky.social! The FMI is a very special place to start your lab: a vibrant international institute with world-class research groups and facilities, core funding, fantastic trainees, great colleagues😅, and a collegial culture. Get in touch if you're interested!
🚨 We're hiring, please share! The FMI seeks a tenure-track Group Leader (Assistant Prof) in Structural Biology 🔬
Innovative scientists in genome regulation, RNA metabolism, or protein homeostasis—especially using cutting-edge approaches—apply now at www.fmi.ch/education-ca...
Innovative scientists in genome regulation, RNA metabolism, or protein homeostasis—especially using cutting-edge approaches—apply now at www.fmi.ch/education-ca...

September 8, 2025 at 8:41 AM
Become our colleague at @fmiscience.bsky.social! The FMI is a very special place to start your lab: a vibrant international institute with world-class research groups and facilities, core funding, fantastic trainees, great colleagues😅, and a collegial culture. Get in touch if you're interested!
Reposted by Luca Giorgetti lab @FMI
This is not a HiC map! Ever wondered if multiple enhancers get activated simultaneously? We measured chromatin accessibility on thousands of molecules by nanopore to create genome-wide co-accessibility maps. Proud of @mathias-boulanger.bsky.social @kasitc.bsky.social Biology in the thread👇

August 18, 2025 at 1:00 PM
This is not a HiC map! Ever wondered if multiple enhancers get activated simultaneously? We measured chromatin accessibility on thousands of molecules by nanopore to create genome-wide co-accessibility maps. Proud of @mathias-boulanger.bsky.social @kasitc.bsky.social Biology in the thread👇
Reposted by Luca Giorgetti lab @FMI
New preprint with @gfudenberg.bsky.social
We find the rate of cohesin loop extrusion in cells is set by NIPBL dosage and tunes many aspects of chromosome folding.
This provides a molecular basis for NIPBL haploinsufficiency in humans. 🧵👇
www.biorxiv.org/content/10.1...
We find the rate of cohesin loop extrusion in cells is set by NIPBL dosage and tunes many aspects of chromosome folding.
This provides a molecular basis for NIPBL haploinsufficiency in humans. 🧵👇
www.biorxiv.org/content/10.1...

NIPBL dosage shapes genome folding by tuning the rate of cohesin loop extrusion
Cohesin loop extrusion is a major driver of chromosome folding, but how its dynamics are controlled to shape the genome remains elusive. Here we disentangle the contributions of the cohesin cofactors ...
www.biorxiv.org
August 16, 2025 at 3:03 AM
New preprint with @gfudenberg.bsky.social
We find the rate of cohesin loop extrusion in cells is set by NIPBL dosage and tunes many aspects of chromosome folding.
This provides a molecular basis for NIPBL haploinsufficiency in humans. 🧵👇
www.biorxiv.org/content/10.1...
We find the rate of cohesin loop extrusion in cells is set by NIPBL dosage and tunes many aspects of chromosome folding.
This provides a molecular basis for NIPBL haploinsufficiency in humans. 🧵👇
www.biorxiv.org/content/10.1...
Reposted by Luca Giorgetti lab @FMI
We are hiring a scientist to lead our 'Build' team at the Seattle Hub for Synthetic Biology. @alleninstitute.org
Job posting below:
alleninstitute.org/careers/jobs...
Please reach out if you have any questions!
Job posting below:
alleninstitute.org/careers/jobs...
Please reach out if you have any questions!

Jobs
We are working to solve the biggest mysteries in bioscience.
alleninstitute.org
August 13, 2025 at 7:03 PM
We are hiring a scientist to lead our 'Build' team at the Seattle Hub for Synthetic Biology. @alleninstitute.org
Job posting below:
alleninstitute.org/careers/jobs...
Please reach out if you have any questions!
Job posting below:
alleninstitute.org/careers/jobs...
Please reach out if you have any questions!
Reposted by Luca Giorgetti lab @FMI
Excited to share the latest work from the lab led by @eharo84.bsky.social, in which we have used synthetic biology to explore the mechanisms by which different types of long-range enhancers ensure robust and precise developmental gene expression
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

Synthetic engineering demonstrates that synergy among enhancers involves an increase in transcriptionally productive enhancer-gene contacts
Enhancers are non-coding cis-regulatory elements that control the expression of distally located genes in a tissue- and time-specific manner. Recent studies indicate that enhancers can differ in their...
www.biorxiv.org
August 11, 2025 at 2:56 PM
Excited to share the latest work from the lab led by @eharo84.bsky.social, in which we have used synthetic biology to explore the mechanisms by which different types of long-range enhancers ensure robust and precise developmental gene expression
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Reposted by Luca Giorgetti lab @FMI
We found a new asymmetry in the large-scale chromosome structure: sister chromatids are systematically shifted by hundreds of kb in the 5′→3′ direction of their inherited strands! The work was led by Flavia Corsi, in close collaboration with the Daniel Gerlich lab.
www.biorxiv.org/content/10.1...
1/
www.biorxiv.org/content/10.1...
1/
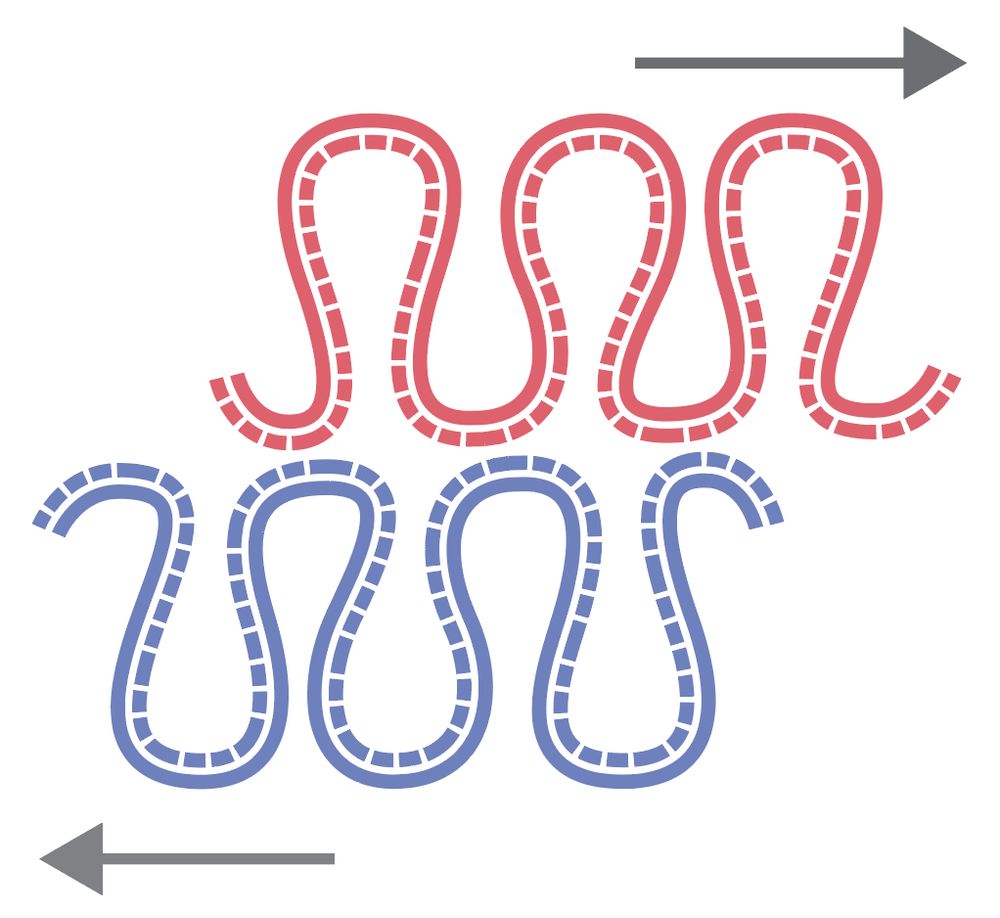
July 15, 2025 at 8:11 AM
We found a new asymmetry in the large-scale chromosome structure: sister chromatids are systematically shifted by hundreds of kb in the 5′→3′ direction of their inherited strands! The work was led by Flavia Corsi, in close collaboration with the Daniel Gerlich lab.
www.biorxiv.org/content/10.1...
1/
www.biorxiv.org/content/10.1...
1/
Reposted by Luca Giorgetti lab @FMI
Our paper describing the Range Extender element which is required and sufficient for long-range enhancer activation at the Shh locus is now available at @nature.com. Congrats to @gracebower.bsky.social who led the study. Below is a brief summary of the main findings www.nature.com/articles/s41... 1/

Range extender mediates long-distance enhancer activity - Nature
The REX element is associated with long-range enhancer–promoter interactions.
www.nature.com
July 2, 2025 at 4:17 PM
Our paper describing the Range Extender element which is required and sufficient for long-range enhancer activation at the Shh locus is now available at @nature.com. Congrats to @gracebower.bsky.social who led the study. Below is a brief summary of the main findings www.nature.com/articles/s41... 1/
Reposted by Luca Giorgetti lab @FMI
Finally out! 🥳 Our paper showing how a transposable element (TE) insertion can cause developmental phenotypes is now published @natgenet.nature.com 🧬🦠🐁
Below is a brief description of the major findings. Check the full version of the paper for more details: www.nature.com/articles/s41588-025-02248-5
Below is a brief description of the major findings. Check the full version of the paper for more details: www.nature.com/articles/s41588-025-02248-5
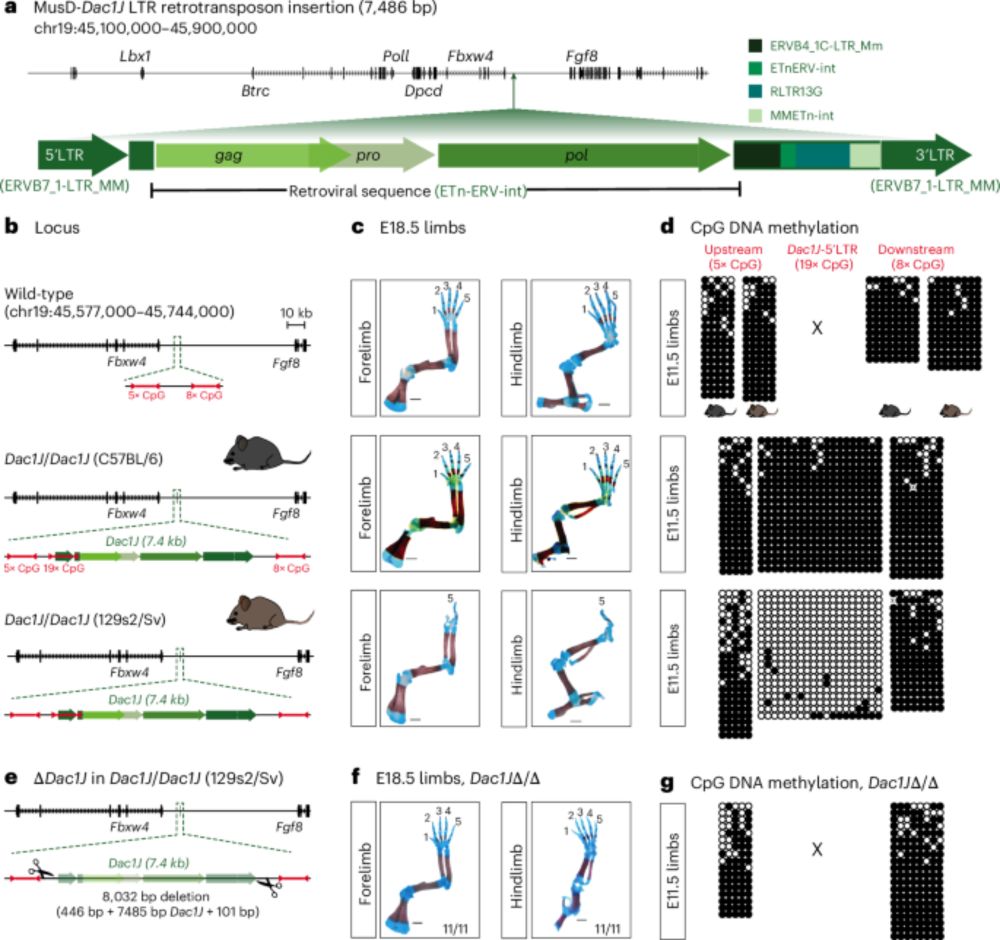
Enhancer adoption by an LTR retrotransposon generates viral-like particles, causing developmental limb phenotypes - Nature Genetics
Activation of an LTR retrotransposon inserted upstream of the Fgf8 gene produces viral-like particles in the mouse developing limb, triggering apoptosis and causing limb malformation. This phenotype c...
www.nature.com
July 9, 2025 at 10:05 AM
Finally out! 🥳 Our paper showing how a transposable element (TE) insertion can cause developmental phenotypes is now published @natgenet.nature.com 🧬🦠🐁
Below is a brief description of the major findings. Check the full version of the paper for more details: www.nature.com/articles/s41588-025-02248-5
Below is a brief description of the major findings. Check the full version of the paper for more details: www.nature.com/articles/s41588-025-02248-5
Reposted by Luca Giorgetti lab @FMI
Our work bridging enhancer-promoter proximity to phenotypic outcomes in vivo is out! Shout out to @olimpiabompadre.bsky.social, to Marie Kmita's lab, and to all the co-authors. www.nature.com/articles/s41...
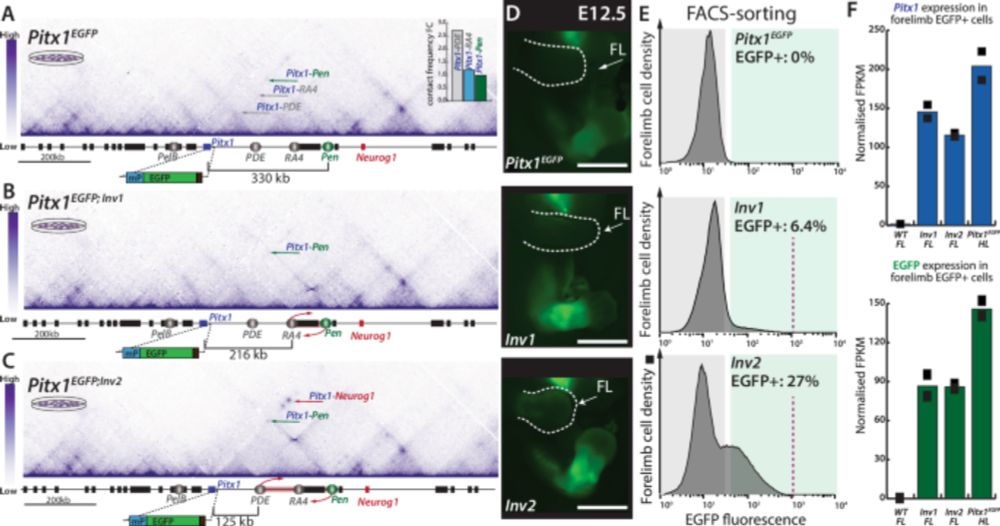
Liebenberg syndrome severity arises from variations in Pitx1 locus topology and proportion of ectopically transcribing cells - Nature Communications
Here the authors show that reducing enhancer-promoter distance at the Pitx1 locus increases proportion of Pitx1 forelimb expressing cells, worsening skeletal defects in Liebenberg syndrome. They also ...
www.nature.com
July 9, 2025 at 11:58 AM
Our work bridging enhancer-promoter proximity to phenotypic outcomes in vivo is out! Shout out to @olimpiabompadre.bsky.social, to Marie Kmita's lab, and to all the co-authors. www.nature.com/articles/s41...
We are looking for a research assistant to join our lab and support ongoing projects with genome engineering in mouse embryonic stem cells! Please get in touch if you are interested.
www.fmi.ch/education-ca...
www.fmi.ch/education-ca...
Friedrich Miescher Institute, open position
Friedrich Miescher Institute, open position
www.fmi.ch
June 26, 2025 at 3:04 PM
We are looking for a research assistant to join our lab and support ongoing projects with genome engineering in mouse embryonic stem cells! Please get in touch if you are interested.
www.fmi.ch/education-ca...
www.fmi.ch/education-ca...
Reposted by Luca Giorgetti lab @FMI
How to find Evolutionary Conserved Enhancers in 2025? 🐣-🐭
Check out our paper - fresh off the press!!!
We find widespread functional conservation of enhancers in absence of sequence homology
Including: a bioinformatic tool to map sequence-diverged enhancers!
rdcu.be/enVDN
github.com/tobiaszehnde...
Check out our paper - fresh off the press!!!
We find widespread functional conservation of enhancers in absence of sequence homology
Including: a bioinformatic tool to map sequence-diverged enhancers!
rdcu.be/enVDN
github.com/tobiaszehnde...

Conservation of regulatory elements with highly diverged sequences across large evolutionary distances
Nature Genetics - Combining functional genomic data from mouse and chicken with a synteny-based strategy identifies positionally conserved cis-regulatory elements in the absence of direct sequence...
rdcu.be
May 27, 2025 at 12:19 PM
How to find Evolutionary Conserved Enhancers in 2025? 🐣-🐭
Check out our paper - fresh off the press!!!
We find widespread functional conservation of enhancers in absence of sequence homology
Including: a bioinformatic tool to map sequence-diverged enhancers!
rdcu.be/enVDN
github.com/tobiaszehnde...
Check out our paper - fresh off the press!!!
We find widespread functional conservation of enhancers in absence of sequence homology
Including: a bioinformatic tool to map sequence-diverged enhancers!
rdcu.be/enVDN
github.com/tobiaszehnde...

