
rdcu.be/e1bBW

rdcu.be/e1bBW
📝 Abstract submission deadline: February 1st.
🎟️ Registration deadline: February 15th.
Learn more and register 👇
www.hubrecht.eu/hubrecht-sym...

📝 Abstract submission deadline: February 1st.
🎟️ Registration deadline: February 15th.
Learn more and register 👇
www.hubrecht.eu/hubrecht-sym...
Very grateful to @dewitlab.bsky.social and @jopkind.bsky.social and to the many other people who supported me along the way.
I’ll be launching the lab soon, focusing on Chromatin Systems Biology.

Very grateful to @dewitlab.bsky.social and @jopkind.bsky.social and to the many other people who supported me along the way.
I’ll be launching the lab soon, focusing on Chromatin Systems Biology.
Come join us at the @sangerinstitute.bsky.social
Sanger is core-funded so you can generate data at scale to train the next generation of models and understanding. Design/Engineering/Chemistry/Proteins/Pathways!
pls RT
tinyurl.com/GenGenFaculty
Come join us at the @sangerinstitute.bsky.social
Sanger is core-funded so you can generate data at scale to train the next generation of models and understanding. Design/Engineering/Chemistry/Proteins/Pathways!
pls RT
tinyurl.com/GenGenFaculty
🧬 Very excited to share our work on dissecting imputation strategies for single-cell histone modification data! academic.oup.com/nargab/artic...
A huge thank you to @robinhweide.bsky.social, @jopkind.bsky.social and Jeroen de Ridder for their collaboration in this project!

🧬 Very excited to share our work on dissecting imputation strategies for single-cell histone modification data! academic.oup.com/nargab/artic...
A huge thank you to @robinhweide.bsky.social, @jopkind.bsky.social and Jeroen de Ridder for their collaboration in this project!
Single-cell Dam&T-seq during mouse corticogenesis reveals how genome–lamina interactions regulate long neuronal genes.
doi.org/10.1101/2025...
#Neurodevelopment #Epigenetics @hubrechtinstitute.bsky.social @oncodeinstitute.bsky.social

Single-cell Dam&T-seq during mouse corticogenesis reveals how genome–lamina interactions regulate long neuronal genes.
doi.org/10.1101/2025...
#Neurodevelopment #Epigenetics @hubrechtinstitute.bsky.social @oncodeinstitute.bsky.social

The full list can be found here: http://www.technologyreview.com/innovators-under-35/2025/.




We find the rate of cohesin loop extrusion in cells is set by NIPBL dosage and tunes many aspects of chromosome folding.
This provides a molecular basis for NIPBL haploinsufficiency in humans. 🧵👇
www.biorxiv.org/content/10.1...

We find the rate of cohesin loop extrusion in cells is set by NIPBL dosage and tunes many aspects of chromosome folding.
This provides a molecular basis for NIPBL haploinsufficiency in humans. 🧵👇
www.biorxiv.org/content/10.1...


Join a vibrant, interdisciplinary community where collaboration and innovation are nurtured at all levels.
Take a look at these four open positions 👇

Join a vibrant, interdisciplinary community where collaboration and innovation are nurtured at all levels.
Take a look at these four open positions 👇
www.biorxiv.org/content/10.1...
1/
www.biorxiv.org/content/10.1...
1/
✉️ The Hubrecht Institute now has its very own PhD program! Applications are open and will be accepted until September 15th. Read more here 👉 www.hubrecht.eu/hipp/

✉️ The Hubrecht Institute now has its very own PhD program! Applications are open and will be accepted until September 15th. Read more here 👉 www.hubrecht.eu/hipp/
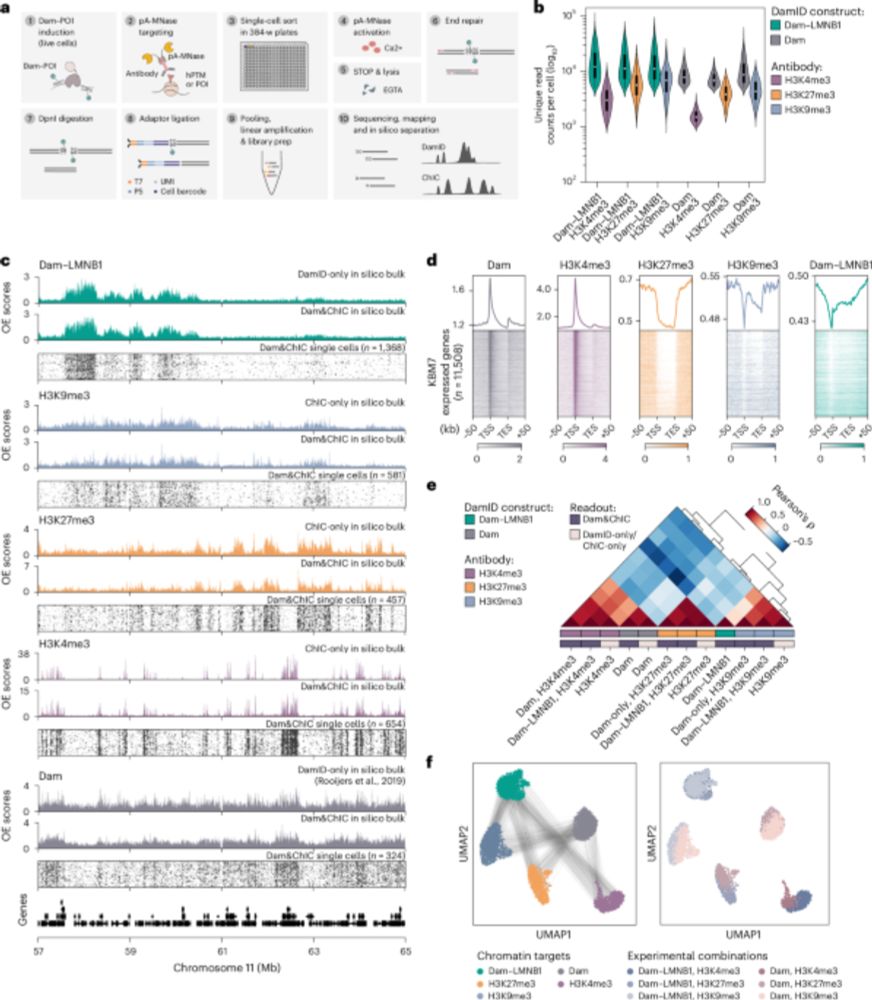
Researchers from the Kind and Van Oudenaarden groups have found a way to study these types of questions: www.nature.com/articles/s41.... Read more below! 👇
@jopkind.bsky.social

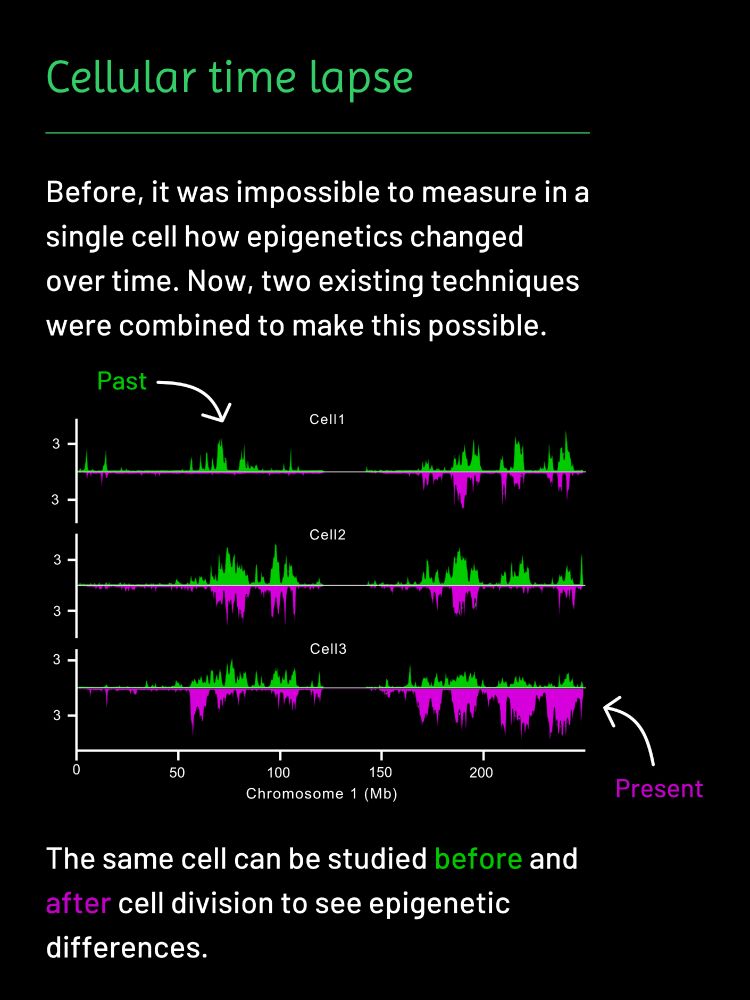
Researchers from the Kind and Van Oudenaarden groups have found a way to study these types of questions: www.nature.com/articles/s41.... Read more below! 👇
@jopkind.bsky.social
www.biorxiv.org/content/10.1...
2/9
www.biorxiv.org/content/10.1...
2/9
7/9

7/9
this is part of a MSCA Doctoral Network on the topic of metabolic regulation of genome function and cell identity.
A EU funded generous fellowship is offered with possibility of industry experience. Apply here:

www.nature.com/articles/s41...
A joy to work with super team Sarah Plattner, Yuka Sugiura, Francisco Falcon and Elly Tanaka.
🧵1/14

www.nature.com/articles/s41...
A joy to work with super team Sarah Plattner, Yuka Sugiura, Francisco Falcon and Elly Tanaka.
🧵1/14

Sharing about 80% of heart disease–causing genes with humans, the zebrafish is an excellent model to study how hearts develop, become diseased and might regenerate.
Sharing about 80% of heart disease–causing genes with humans, the zebrafish is an excellent model to study how hearts develop, become diseased and might regenerate.
Ever wondered how cells prepare their genomes to enable new cell-fates? In this team up with the Kind lab, we show that genes are repositioned in the nucleus to get ready for future activation and tissue formation. Read 🧵👇 to find out how and when this happens!
doi.org/10.1101/2025...

Ever wondered how cells prepare their genomes to enable new cell-fates? In this team up with the Kind lab, we show that genes are repositioned in the nucleus to get ready for future activation and tissue formation. Read 🧵👇 to find out how and when this happens!
doi.org/10.1101/2025...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

