Head of Generative Genomics, Wellcome Sanger Institute, Cambridge, UK
Systems + Synthetic Biology, CRG, Barcelona
http://barcelonacollaboratorium.com http://allox.bio https://www.sanger.ac.uk/programme/generative-and-synthetic-genomics/
Come join us at the @sangerinstitute.bsky.social
Sanger is core-funded so you can generate data at scale to train the next generation of models and understanding. Design/Engineering/Chemistry/Proteins/Pathways!
pls RT
tinyurl.com/GenGenFaculty
Come join us at the @sangerinstitute.bsky.social
Sanger is core-funded so you can generate data at scale to train the next generation of models and understanding. Design/Engineering/Chemistry/Proteins/Pathways!
pls RT
tinyurl.com/GenGenFaculty
Are Transcription Factors really 'undruggable'?
www.biorxiv.org/content/10.1...

Are Transcription Factors really 'undruggable'?
www.biorxiv.org/content/10.1...
Full deep mutational scan of an entire human protein interaction domain family
2M quantitative measurement of protein-protein interaction -> deep learning model to predict PPI from sequence
More details 👇
We used ddPCA to map the genetic architecture of the entire human bZIP interaction network.
www.biorxiv.org/content/10.1...
Thanks to all our co-authors for the great collaboration!

Full deep mutational scan of an entire human protein interaction domain family
2M quantitative measurement of protein-protein interaction -> deep learning model to predict PPI from sequence
More details 👇

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
🧬 By charting a protein’s sequence universe, we could rationalize which versions were kept through evolution – and why many stable ones were not.
🧬 By charting a protein’s sequence universe, we could rationalize which versions were kept through evolution – and why many stable ones were not.
@xt117.bsky.social
biorxiv.org/content/10.1...

@xt117.bsky.social
biorxiv.org/content/10.1...
www.sanger.ac.uk

www.sanger.ac.uk
Aina's new preprint containing seven complete comparative allosteric maps biorxiv.org/content/10.1...
Aina's new preprint containing seven complete comparative allosteric maps biorxiv.org/content/10.1...
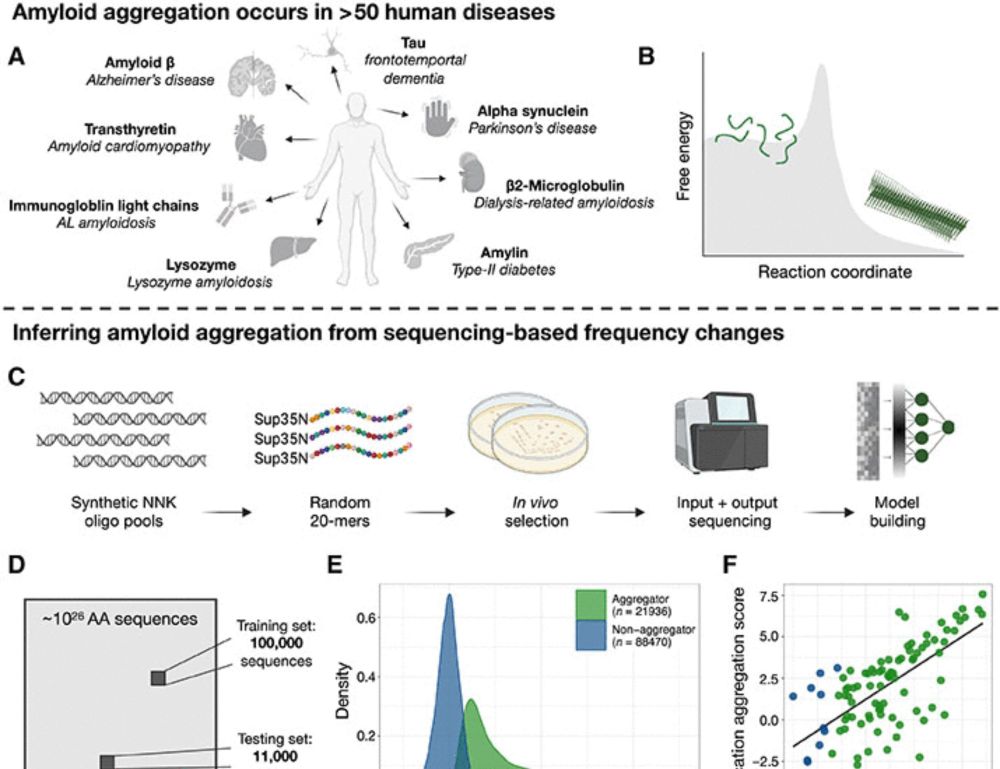
In our new study we cracked open the black box of early protein aggregation, and the findings could reshape how we fight neurodegeneration. 🧵👇
www.science.org/doi/10.1126/...
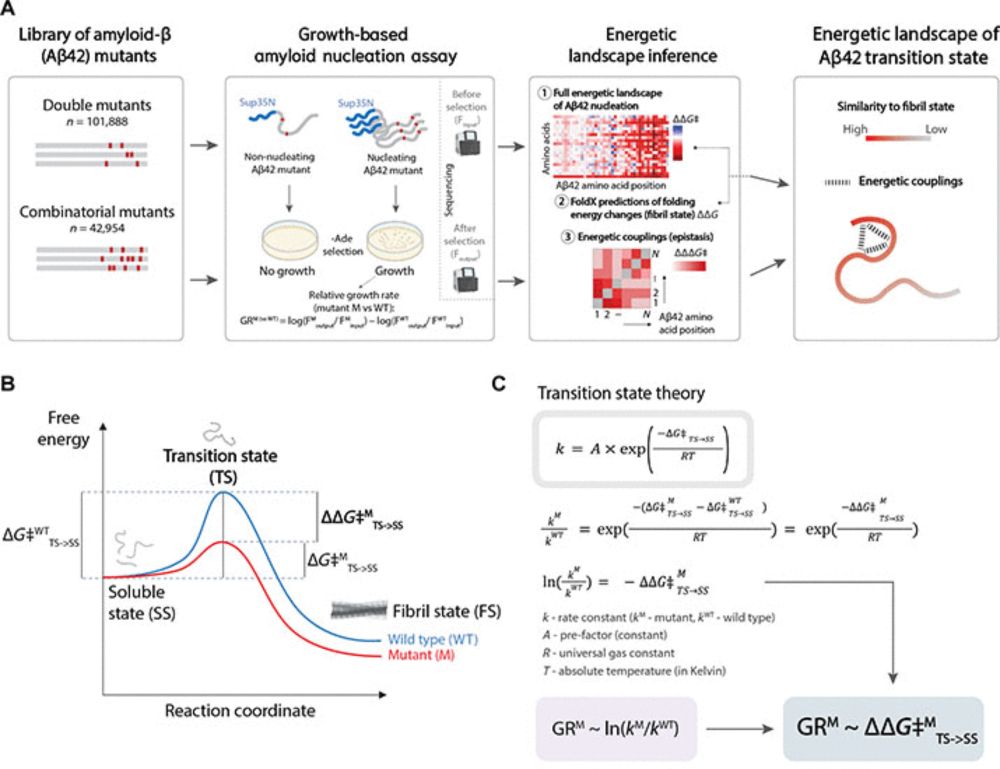
In our new study we cracked open the black box of early protein aggregation, and the findings could reshape how we fight neurodegeneration. 🧵👇
www.science.org/doi/10.1126/...
3,500 mutants, to single-site saturation 🧬
>45,000 binding and abundance measurements 📶
Very happy to present our latest work – where deep mutational scanning meets the world of small molecules.
www.biorxiv.org/content/10.1...
With @benlehner.bsky.social
[1/7]

3,500 mutants, to single-site saturation 🧬
>45,000 binding and abundance measurements 📶
Very happy to present our latest work – where deep mutational scanning meets the world of small molecules.
www.biorxiv.org/content/10.1...
With @benlehner.bsky.social
[1/7]

A new platform for high resolution functional mapping of GPCRs with massive mutagenesis. We identify the core activation network, residues involved in biased signaling, and generate >7,000 full dose response curves. With @benlehner.bsky.social

A new platform for high resolution functional mapping of GPCRs with massive mutagenesis. We identify the core activation network, residues involved in biased signaling, and generate >7,000 full dose response curves. With @benlehner.bsky.social
www.biorxiv.org/content/10.1...

Building AI models or the data to train them?
Core funding of >$130M a year for a faculty of ~30.
www.nature.com/naturecareer...
acrobat.adobe.com/id/urn:aaid:...
pls RT!

Building AI models or the data to train them?
Core funding of >$130M a year for a faculty of ~30.
www.nature.com/naturecareer...
acrobat.adobe.com/id/urn:aaid:...
pls RT!
Organised by @bennibolo.bsky.social @jonnyfrazer.bsky.social @benlehner.bsky.social @muffley.bsky.social @muffley.bsky.social and Mafalda Dias.
More info: events.ibecbarcelona.eu/mutational-s...

Organised by @bennibolo.bsky.social @jonnyfrazer.bsky.social @benlehner.bsky.social @muffley.bsky.social @muffley.bsky.social and Mafalda Dias.
More info: events.ibecbarcelona.eu/mutational-s...
rdcu.be/d8xLv (1/6)
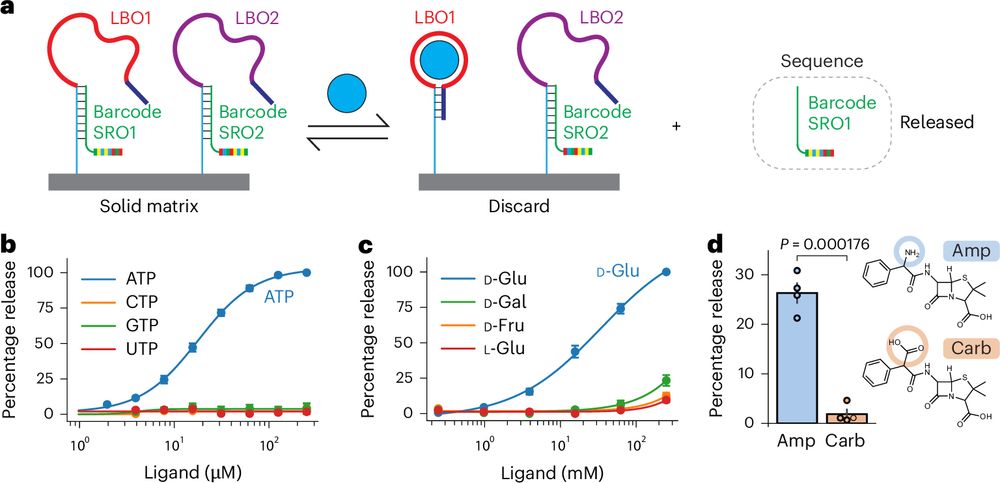
rdcu.be/d8xLv (1/6)
📌Structural Bioinformatics Scientist 👉 tinyurl.com/3rs46j4t
📌Platform Bioinformatician 👉 tinyurl.com/nyvazbke
📌Computational Chemist 👉 tinyurl.com/5n6vpem3
Apply or repost to help us spread the word 🙏

📌Structural Bioinformatics Scientist 👉 tinyurl.com/3rs46j4t
📌Platform Bioinformatician 👉 tinyurl.com/nyvazbke
📌Computational Chemist 👉 tinyurl.com/5n6vpem3
Apply or repost to help us spread the word 🙏

📌Structural Bioinformatics Scientist 👉 tinyurl.com/3rs46j4t
📌Platform Bioinformatician 👉 tinyurl.com/nyvazbke
📌Computational Chemist 👉 tinyurl.com/5n6vpem3
Apply or repost to help us spread the word 🙏
The CRG researchers are among a select group of 134 scientists across Europe who will explore the commercial potential of their research. #ERCPoC



