Human and viral RNAs.
Hubrecht Institute, Oncode Institute and dept. of Bionanoscience, TU Delft
rdcu.be/e1bBW

rdcu.be/e1bBW
www.biorxiv.org/content/10.6...

www.biorxiv.org/content/10.6...
www.biorxiv.org/content/10.6...
www.biorxiv.org/content/10.6...
A new methodology developed in our group, spearheaded by Jasper van den Ende, provides insight in this process.
EV-FUSIM: an EV-Fusion Spatiotemporal Imaging Method.
www.biorxiv.org/content/10.1...

A new methodology developed in our group, spearheaded by Jasper van den Ende, provides insight in this process.
EV-FUSIM: an EV-Fusion Spatiotemporal Imaging Method.
www.biorxiv.org/content/10.1...
✉️ The Hubrecht Institute now has its very own PhD program! Applications are open and will be accepted until September 15th. Read more here 👉 www.hubrecht.eu/hipp/

Prkra dimer senses double-stranded RNAs to dictate global translation efficiency: Molecular Cell www.cell.com/molecular-ce...

Prkra dimer senses double-stranded RNAs to dictate global translation efficiency: Molecular Cell www.cell.com/molecular-ce...
The HTP aims to promote scientific excellence in the Netherlands by supporting talented minority students in pursuing a career in scientific research.
Read more in the flyer and on www.hubrecht.eu/about-us/hub...


The HTP aims to promote scientific excellence in the Netherlands by supporting talented minority students in pursuing a career in scientific research.
Read more in the flyer and on www.hubrecht.eu/about-us/hub...
www.ammodo-science.org/researches/m...

www.ammodo-science.org/researches/m...
doi.org/10.1101/2025...

doi.org/10.1101/2025...

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
www.cell.com/cell/fulltex...
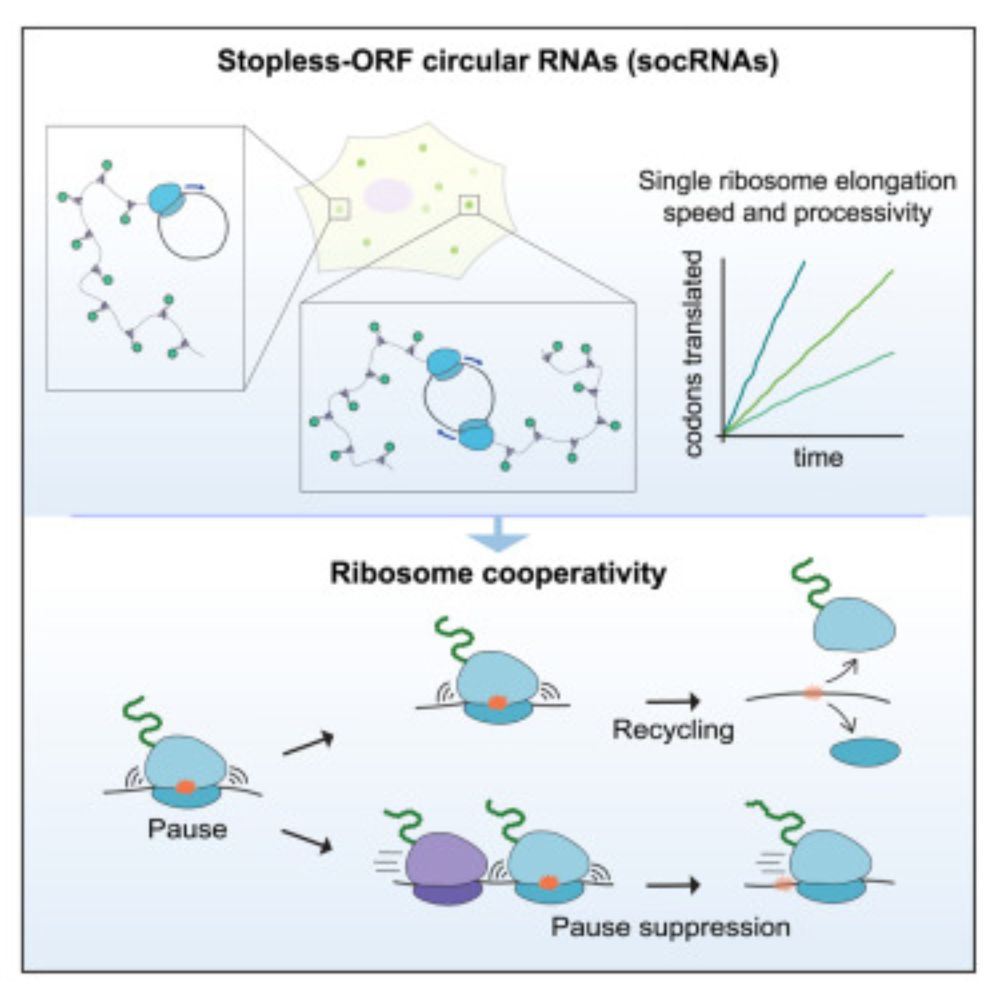
www.cell.com/cell/fulltex...
https://go.nature.com/4hDtGhL

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...


