www.biorxiv.org/content/10.6...

www.biorxiv.org/content/10.6...
www.biorxiv.org/content/10.6...
www.biorxiv.org/content/10.6...
www.cell.com/cell/fulltex...
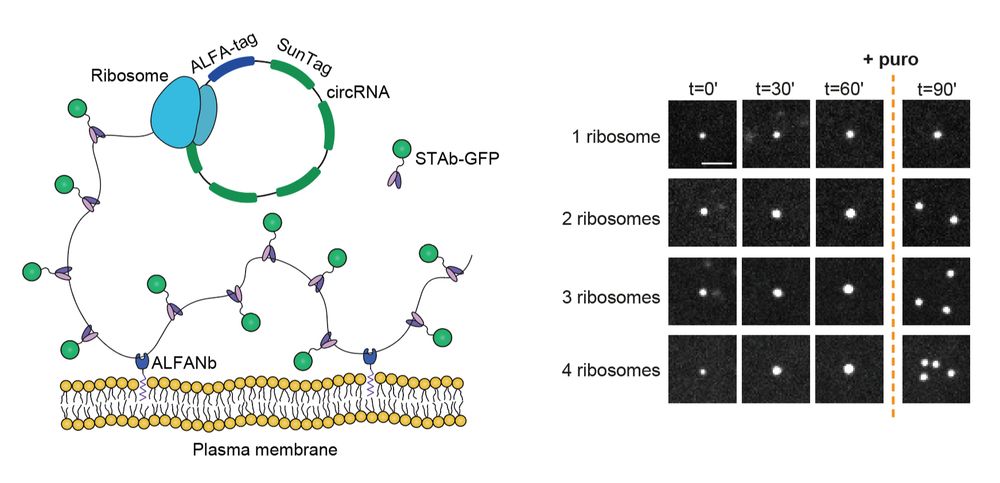
www.cell.com/cell/fulltex...
www.cell.com/cell/fulltex...
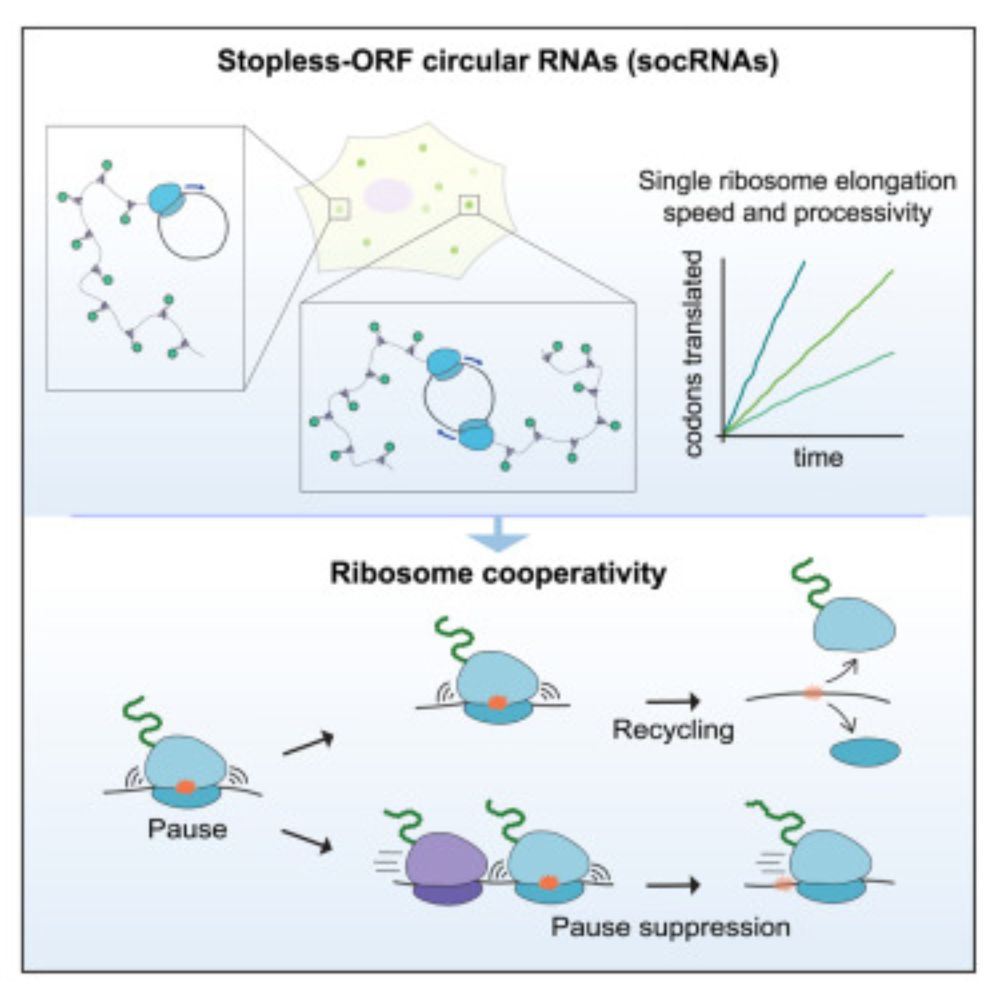
www.cell.com/cell/fulltex...

