Luca Giorgetti lab @FMI
@lucagiorgetti.bsky.social
We study transcriptional regulation and chromosome folding using an interdisciplinary approach combining wet- and dry-lab methods.
https://giorgettilab.org
@fmiscience.bsky.social
https://giorgettilab.org
@fmiscience.bsky.social
17/ ... and nicely predicts @elphegenoralab.bsky.social @karissalhansen.bsky.social's data at the Car2 locus www.biorxiv.org/content/10.1..., using changes in cohesin occupancy and extrusion velocity measured with @gfudenberg.bsky.social in www.biorxiv.org/content/10.1...:

September 24, 2025 at 9:45 PM
17/ ... and nicely predicts @elphegenoralab.bsky.social @karissalhansen.bsky.social's data at the Car2 locus www.biorxiv.org/content/10.1..., using changes in cohesin occupancy and extrusion velocity measured with @gfudenberg.bsky.social in www.biorxiv.org/content/10.1...:
15/ This happens because by selecting longer and longer encounters, one selects more and more loop-extrusion-driven events, the probability of which is exponential as a function of genomic distance!

September 24, 2025 at 9:45 PM
15/ This happens because by selecting longer and longer encounters, one selects more and more loop-extrusion-driven events, the probability of which is exponential as a function of genomic distance!
14/ This simple model also predicts that transcription levels should decrease exponentially as a function of genomic distance between an enhancer and promoter, exactly as we verified using data from our previous work!

September 24, 2025 at 9:45 PM
14/ This simple model also predicts that transcription levels should decrease exponentially as a function of genomic distance between an enhancer and promoter, exactly as we verified using data from our previous work!
13/ Strikingly, this simple hypothesis predicts that: 1) average transcription levels should increase nonlinearly as a function of enhancer-promoter contact probabilities, as we and others previously observed: www.nature.com/articles/s41... and also work from e.g. Boettiger, de Laat, Wysocka labs

September 24, 2025 at 9:45 PM
13/ Strikingly, this simple hypothesis predicts that: 1) average transcription levels should increase nonlinearly as a function of enhancer-promoter contact probabilities, as we and others previously observed: www.nature.com/articles/s41... and also work from e.g. Boettiger, de Laat, Wysocka labs
12/...so the probability that such events result in transcription should thus increase when increasing the time that an enhancer and promoter spend in physical proximity. We thus asked: What if only encounters whose duration exceeds an arbitrary cutoff time are productive for transcription?

September 24, 2025 at 9:45 PM
12/...so the probability that such events result in transcription should thus increase when increasing the time that an enhancer and promoter spend in physical proximity. We thus asked: What if only encounters whose duration exceeds an arbitrary cutoff time are productive for transcription?
10/ …and managed to film cells in 3D every 2s with an error in the xy plan of only ~30nm, many times better than our previous measurements! With this in hand, @nessim and @mattia were able to clearly distinguish the appearance of long-lived encounters exclusively in the presence of cohesin!


September 24, 2025 at 9:45 PM
10/ …and managed to film cells in 3D every 2s with an error in the xy plan of only ~30nm, many times better than our previous measurements! With this in hand, @nessim and @mattia were able to clearly distinguish the appearance of long-lived encounters exclusively in the presence of cohesin!
8/We then asked if we could detect extrusion-driven encounters in living cells. We resorted to a cell line previously established in the lab by @piamach.bsky.social, allowing to measure the distance between two operator arrays separated by 150kb in cells mESC where RAD21 can be inducibly degraded.

September 24, 2025 at 9:45 PM
8/We then asked if we could detect extrusion-driven encounters in living cells. We resorted to a cell line previously established in the lab by @piamach.bsky.social, allowing to measure the distance between two operator arrays separated by 150kb in cells mESC where RAD21 can be inducibly degraded.
6/ We showed that these encounters only arise when cohesin loads approximately midway between two loci, extrudes them into the encounter radius, and pushes them out after some time.

September 24, 2025 at 9:45 PM
6/ We showed that these encounters only arise when cohesin loads approximately midway between two loci, extrudes them into the encounter radius, and pushes them out after some time.
5/Mattia also discovered that loop extrusion creates a new class of encounters which are extremely rare, but last substantially longer than random polymer collisions. The duration of such encounters depends explicitly on the velocity of extrusion, and it thus a signature of active extrusion!

September 24, 2025 at 9:45 PM
5/Mattia also discovered that loop extrusion creates a new class of encounters which are extremely rare, but last substantially longer than random polymer collisions. The duration of such encounters depends explicitly on the velocity of extrusion, and it thus a signature of active extrusion!
2/ We started by asking: How often, and for how long, do two genomic sequences meet in the cell nucleus within an arbitrary encounter radius? And how does this depend on the loop extrusion activity of cohesin?

September 24, 2025 at 9:45 PM
2/ We started by asking: How often, and for how long, do two genomic sequences meet in the cell nucleus within an arbitrary encounter radius? And how does this depend on the loop extrusion activity of cohesin?
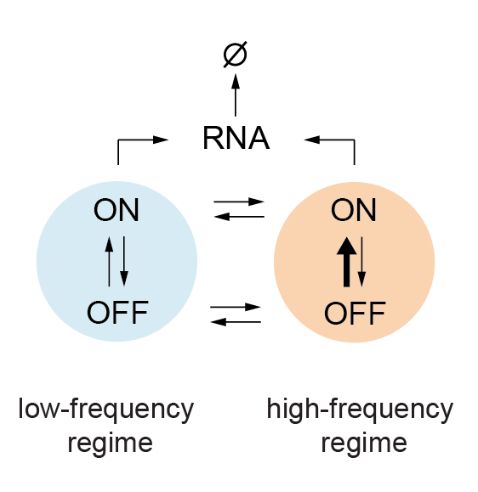
(8/n) This model not only fits the observations on single clones but also correctly predicts the effect of changing the position of the enhancer under the hypothesis that the enhancer only modulates the promoter’s ability to switch from the low- to the high-frequency regime.

March 29, 2025 at 12:46 PM
(8/n) This model not only fits the observations on single clones but also correctly predicts the effect of changing the position of the enhancer under the hypothesis that the enhancer only modulates the promoter’s ability to switch from the low- to the high-frequency regime.
(7/n) … in which the promoter stochastically switches between a ‘basal’ low burst-frequency regime and a high burst-frequency regime where clusters of bursts become more frequent
March 29, 2025 at 12:46 PM
(7/n) … in which the promoter stochastically switches between a ‘basal’ low burst-frequency regime and a high burst-frequency regime where clusters of bursts become more frequent
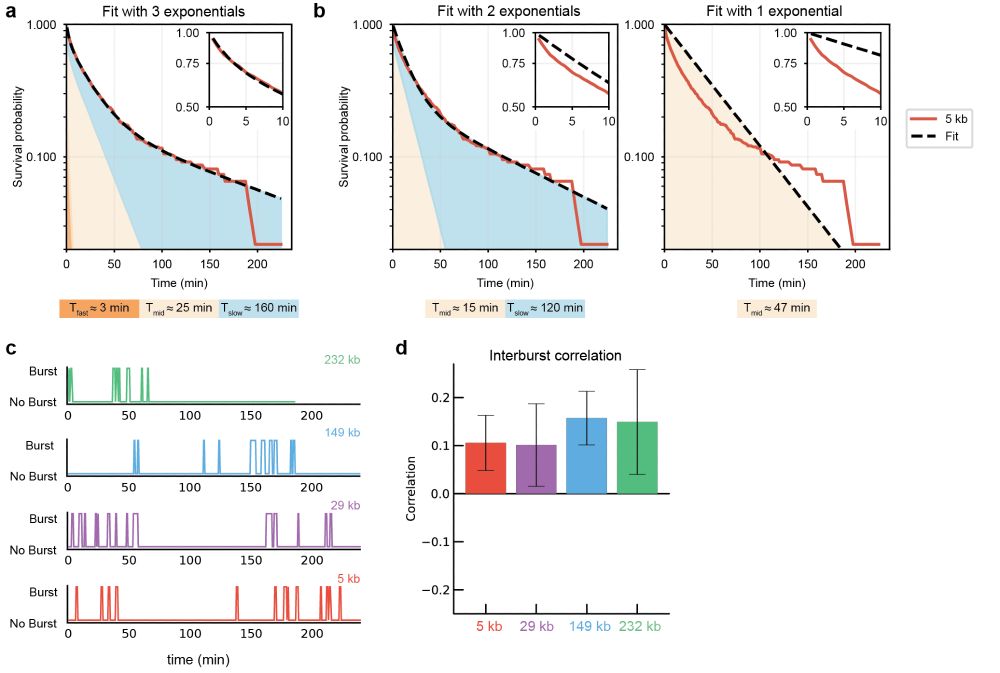
(5/n) We also observed that bursts occur in clusters, with inter-burst duration distributed across three timescales and correlated in time
March 29, 2025 at 12:46 PM
(5/n) We also observed that bursts occur in clusters, with inter-burst duration distributed across three timescales and correlated in time
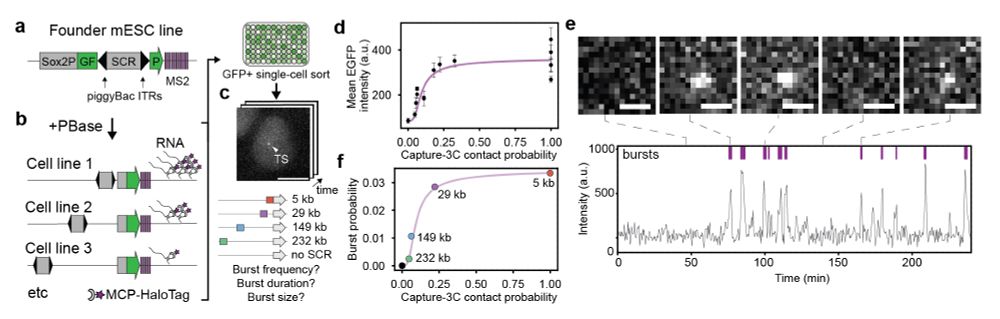
(4/n) When it is located close to the promoter, the enhancer drives more frequent bursts than when it is located far away, resulting in a more uniform transcriptional output. Thus enhancer location within a cis-regulatory landscape is a major determinant of how accurately a gene is transcribed!

March 29, 2025 at 12:46 PM
(4/n) When it is located close to the promoter, the enhancer drives more frequent bursts than when it is located far away, resulting in a more uniform transcriptional output. Thus enhancer location within a cis-regulatory landscape is a major determinant of how accurately a gene is transcribed!
(3/n) We show that enhancer position controls the promoter burst frequency, but not burst duration and size

March 29, 2025 at 12:46 PM
(3/n) We show that enhancer position controls the promoter burst frequency, but not burst duration and size
(2/n) We modified our enhancer mobilization assay (www.nature.com/articles/s41...) to enable MS2-mediated visualization of active transcription sites when the SCR enhancer is hopped and reinserted into different random positions around an ectopic copy of the Sox2 promoter inside a ‘neutral’ TAD.
March 29, 2025 at 12:46 PM
(2/n) We modified our enhancer mobilization assay (www.nature.com/articles/s41...) to enable MS2-mediated visualization of active transcription sites when the SCR enhancer is hopped and reinserted into different random positions around an ectopic copy of the Sox2 promoter inside a ‘neutral’ TAD.
Celebrating 10 years of our lab with a new preprint:
www.biorxiv.org/content/10.1...
How does enhancer location within a TAD control transcriptional bursts from a cognate promoter?
Experiments by Jana Tünnermann and modelling by Gregory Roth
www.biorxiv.org/content/10.1...
How does enhancer location within a TAD control transcriptional bursts from a cognate promoter?
Experiments by Jana Tünnermann and modelling by Gregory Roth

March 29, 2025 at 12:46 PM
Celebrating 10 years of our lab with a new preprint:
www.biorxiv.org/content/10.1...
How does enhancer location within a TAD control transcriptional bursts from a cognate promoter?
Experiments by Jana Tünnermann and modelling by Gregory Roth
www.biorxiv.org/content/10.1...
How does enhancer location within a TAD control transcriptional bursts from a cognate promoter?
Experiments by Jana Tünnermann and modelling by Gregory Roth
(8/n) This model not only fits the observations on single clones but also correctly predicts the effect of changing the position of the enhancer under the hypothesis that the enhancer only modulates the promoter’s ability to switch from the low- to the high-frequency regime.

March 29, 2025 at 12:35 PM
(8/n) This model not only fits the observations on single clones but also correctly predicts the effect of changing the position of the enhancer under the hypothesis that the enhancer only modulates the promoter’s ability to switch from the low- to the high-frequency regime.
(7/n) … in which the promoter stochastically switches between a ‘basal’ low burst-frequency regime and a high burst-frequency regime where clusters of bursts become more frequent

March 29, 2025 at 12:35 PM
(7/n) … in which the promoter stochastically switches between a ‘basal’ low burst-frequency regime and a high burst-frequency regime where clusters of bursts become more frequent
(5/n) We also observed that bursts occur in clusters, with inter-burst duration distributed across three timescales and correlated in time

March 29, 2025 at 12:35 PM
(5/n) We also observed that bursts occur in clusters, with inter-burst duration distributed across three timescales and correlated in time
(4/n) When it is located close to the promoter, the enhancer drives more frequent bursts than when it is located far away, resulting in a more uniform transcriptional output. Thus enhancer location within a cis-regulatory landscape is a major determinant of how accurately a gene is transcribed!

March 29, 2025 at 12:35 PM
(4/n) When it is located close to the promoter, the enhancer drives more frequent bursts than when it is located far away, resulting in a more uniform transcriptional output. Thus enhancer location within a cis-regulatory landscape is a major determinant of how accurately a gene is transcribed!
(3/n) We show that enhancer position controls the promoter burst frequency, but not burst duration and size

March 29, 2025 at 12:35 PM
(3/n) We show that enhancer position controls the promoter burst frequency, but not burst duration and size
(2/n) We modified our enhancer mobilization assay (www.nature.com/articles/s41...) to enable MS2-mediated visualization of active transcription sites when the SCR enhancer is mobilized and reinserted into different random positions around an ectopic copy of the Sox2 promoter inside a ‘neutral’ TAD.

March 29, 2025 at 12:35 PM
(2/n) We modified our enhancer mobilization assay (www.nature.com/articles/s41...) to enable MS2-mediated visualization of active transcription sites when the SCR enhancer is mobilized and reinserted into different random positions around an ectopic copy of the Sox2 promoter inside a ‘neutral’ TAD.

