
Website: https://www.unige.ch/medecine/gede/en/research-groups/999andrey
In this new study we wondered about the role of cohesin loading at enhancers for long-range transcriptional control
www.biorxiv.org/content/10.6...
detailed 🧵👇
In this new study we wondered about the role of cohesin loading at enhancers for long-range transcriptional control
www.biorxiv.org/content/10.6...
detailed 🧵👇


#Single-cell genomics, #Epigenomics & #3Dgenome biology in #Zebrafish #DevBio 🐟
📍 @cabd-upo-csic.bsky.social, CABD, Seville (Spain)
🕒 Full-time, 3-year position (start March 2026)
📅 Apply by Feb 15, 2026
👉 drive.google.com/file/d/1M8rp...
#Single-cell genomics, #Epigenomics & #3Dgenome biology in #Zebrafish #DevBio 🐟
📍 @cabd-upo-csic.bsky.social, CABD, Seville (Spain)
🕒 Full-time, 3-year position (start March 2026)
📅 Apply by Feb 15, 2026
👉 drive.google.com/file/d/1M8rp...
1/14
1/14
Led by @raquelrouco.bsky.social, this study establishes a new framework to study how enhancer landscapes act sequentially at developmental loci and are silenced to shape gene expression patterns. (1/n)
Led by @raquelrouco.bsky.social, this study establishes a new framework to study how enhancer landscapes act sequentially at developmental loci and are silenced to shape gene expression patterns. (1/n)

www.biorxiv.org/content/10.6...

We now offer a 4-years PhD contract to investigate whether the dirsuption of this regulatory mechanism can lead to congenital defects. More details 👇

We now offer a 4-years PhD contract to investigate whether the dirsuption of this regulatory mechanism can lead to congenital defects. More details 👇
Editorial @nature.com this week
And 7 basic science discoveries that changed the world
nature.com/articles/d41...
nature.com/articles/d41...


Editorial @nature.com this week
And 7 basic science discoveries that changed the world
nature.com/articles/d41...
nature.com/articles/d41...
We very happy to share our latest work trying to understand enhancer-promoter compatibility.
I am very excited about the results of @blanka-majchrzycka.bsky.social, which changed the way I think about promoters
www.biorxiv.org/content/10.1...

We very happy to share our latest work trying to understand enhancer-promoter compatibility.
I am very excited about the results of @blanka-majchrzycka.bsky.social, which changed the way I think about promoters
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
A🧵👇
www.biorxiv.org/content/10.1...
A🧵👇

I'm sure I've forgotten lots of people, so don't hesitate to let me know so I can add you to the list.
go.bsky.app/8vTgeXB
I'm sure I've forgotten lots of people, so don't hesitate to let me know so I can add you to the list.
go.bsky.app/8vTgeXB
Tissue-specific changes in 3D genome folding cause enhancer hijacking in Liebenberg syndrome
But it’s NOT the overall expression level that drives the phenotype - it’s the proportion of cells hijacked into misexpressing Pitx1 that matters
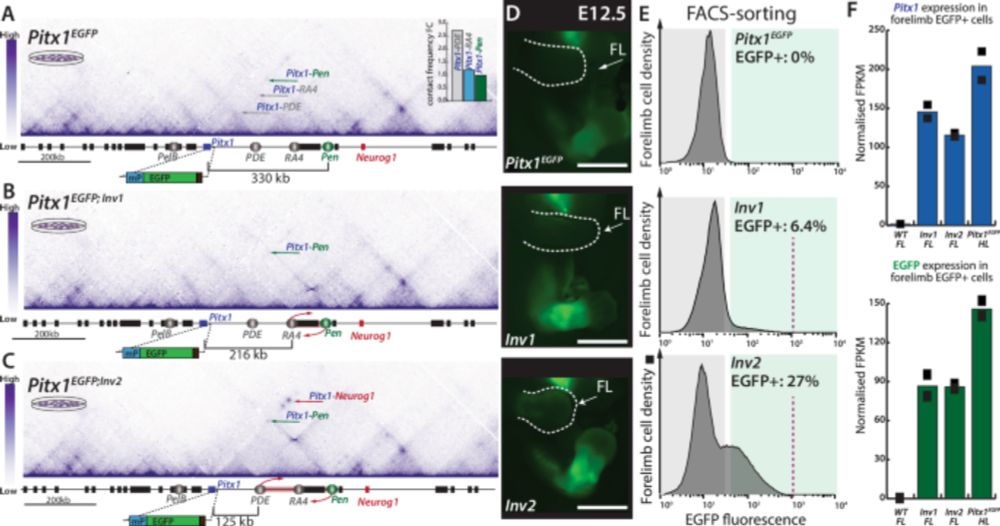
Tissue-specific changes in 3D genome folding cause enhancer hijacking in Liebenberg syndrome
But it’s NOT the overall expression level that drives the phenotype - it’s the proportion of cells hijacked into misexpressing Pitx1 that matters

Below is a brief description of the major findings. Check the full version of the paper for more details: www.nature.com/articles/s41588-025-02248-5
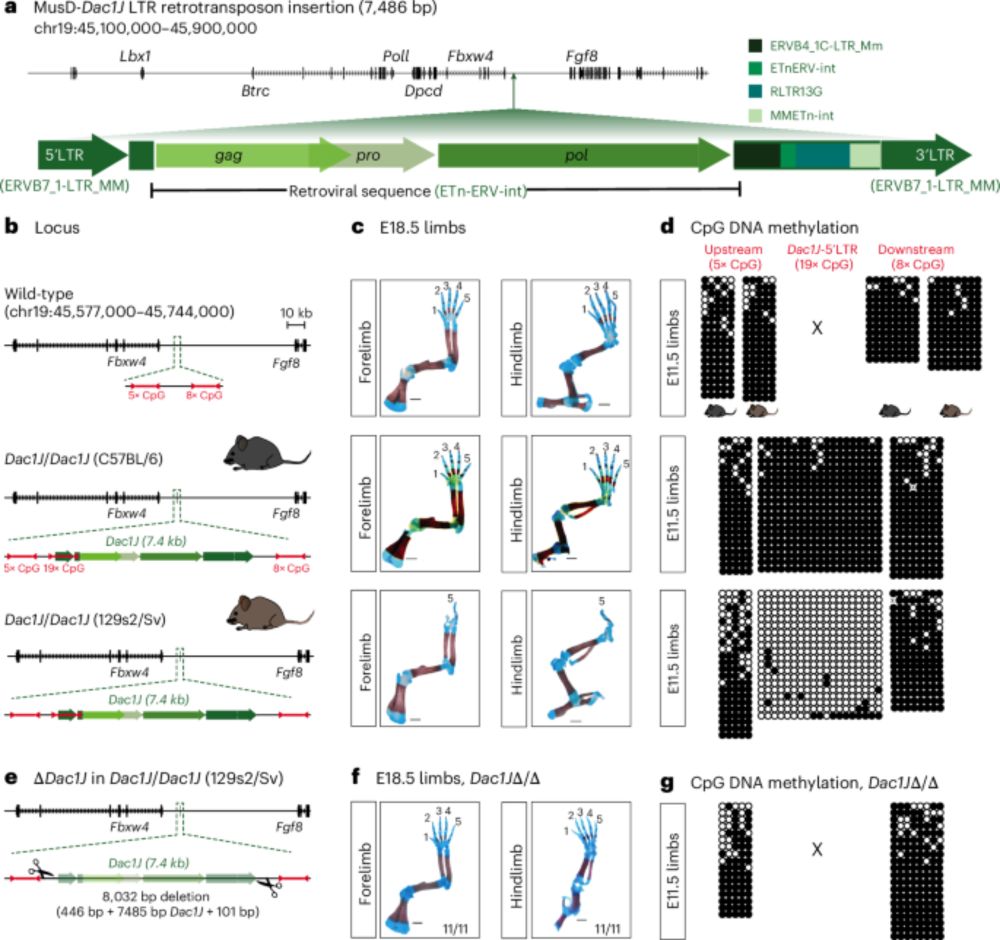
Below is a brief description of the major findings. Check the full version of the paper for more details: www.nature.com/articles/s41588-025-02248-5


Come join our HFSP team to uncover how chromatin moves in cells and what this means for genome function! Great opportunity to combine single-cell genomics, live imaging and polymer physics in the unique mammalian retina with @andersshansen.bsky.social, Davide Michieletto & Sandra Tenreiro

Come join our HFSP team to uncover how chromatin moves in cells and what this means for genome function! Great opportunity to combine single-cell genomics, live imaging and polymer physics in the unique mammalian retina with @andersshansen.bsky.social, Davide Michieletto & Sandra Tenreiro
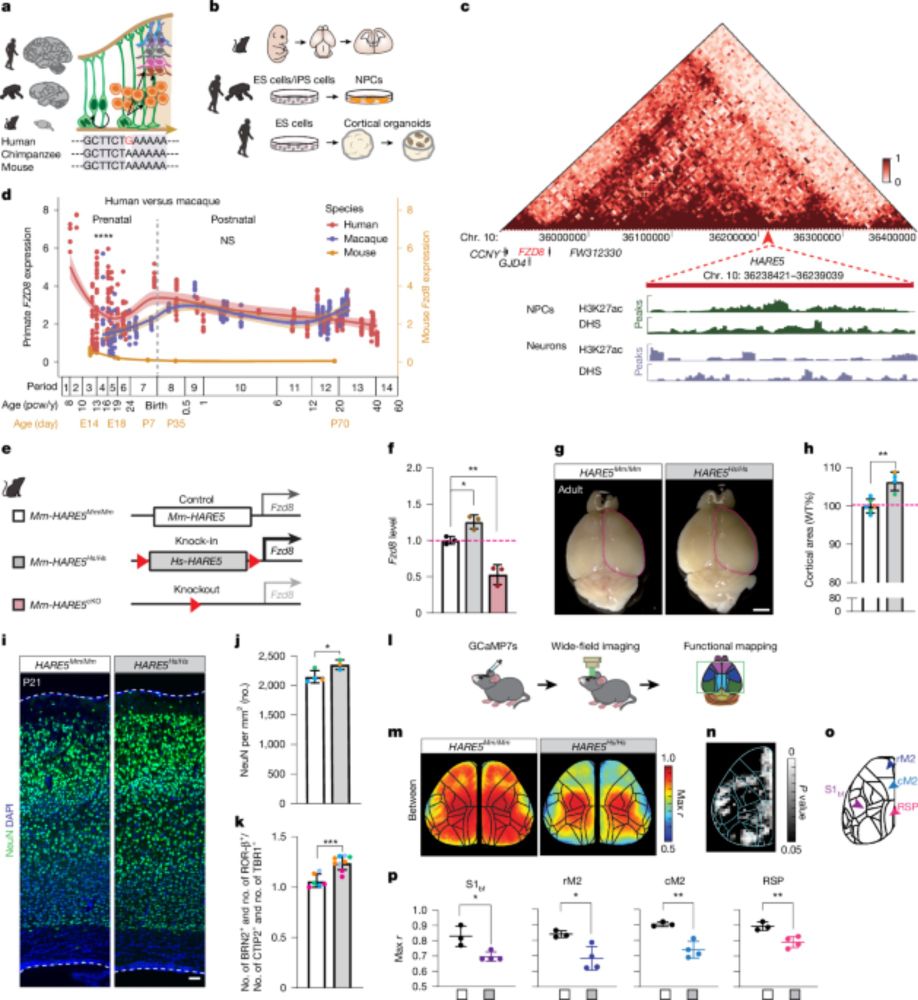
Check out our paper - fresh off the press!!!
We find widespread functional conservation of enhancers in absence of sequence homology
Including: a bioinformatic tool to map sequence-diverged enhancers!
rdcu.be/enVDN
github.com/tobiaszehnde...

Check out our paper - fresh off the press!!!
We find widespread functional conservation of enhancers in absence of sequence homology
Including: a bioinformatic tool to map sequence-diverged enhancers!
rdcu.be/enVDN
github.com/tobiaszehnde...
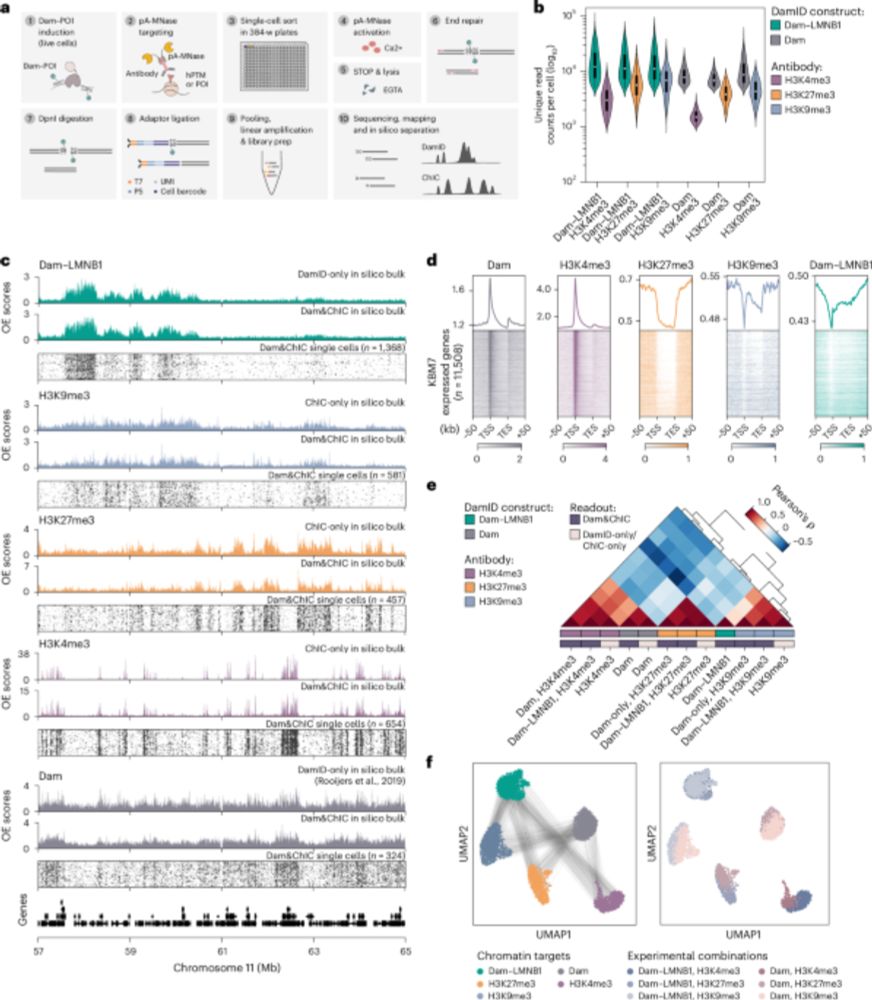
Ever wondered how cells prepare their genomes to enable new cell-fates? In this team up with the Kind lab, we show that genes are repositioned in the nucleus to get ready for future activation and tissue formation. Read 🧵👇 to find out how and when this happens!
doi.org/10.1101/2025...



