
Eliminate your opponents of course.
Recently, my friend @fernpizza.bsky.social showed how plasmids compete intracellularly (check out his paper published in Science today!). With @baym.lol, we now know they can fight.
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Or from a single dish of HEK cell culture in the case of two membrane proteins.
Out in Nature Methods now! lnkd.in/gpyBSceg
Wonderful collaboration with the Efremov lab.

Or from a single dish of HEK cell culture in the case of two membrane proteins.
Out in Nature Methods now! lnkd.in/gpyBSceg
Wonderful collaboration with the Efremov lab.
Meet MetaEdit: a platform for pathway-scale metagenomic editing inside the gut microbiome. science.org/doi/10.1126/...

Meet MetaEdit: a platform for pathway-scale metagenomic editing inside the gut microbiome. science.org/doi/10.1126/...
Brilliant study led by @fmacleod.bsky.social and Andriko von Kügelgen. Tight collaboration with @buzzbaum.bsky.social and lab. Congrats to all authors!
www.biorxiv.org/content/10.1...

Brilliant study led by @fmacleod.bsky.social and Andriko von Kügelgen. Tight collaboration with @buzzbaum.bsky.social and lab. Congrats to all authors!
www.biorxiv.org/content/10.1...
🧬We'll be finding funky new RNA biology, mainly by looking at reverse transcriptases (i.e. the Best Enzymes In The World)🧬
annnd: I'm hiring - come join! Especially postdocs and PhD students - please get in touch (NYC is great)

🧬We'll be finding funky new RNA biology, mainly by looking at reverse transcriptases (i.e. the Best Enzymes In The World)🧬
annnd: I'm hiring - come join! Especially postdocs and PhD students - please get in touch (NYC is great)
Please spread the word!
ista.ac.at/en/job/tenur...

Please spread the word!
ista.ac.at/en/job/tenur...
@roisnehamelinf.bsky.social & co discovered that Wadjet, an SMC complex involved in bacterial DNA immunity, performs some impressive molecular gymnastics 🤸♂️🤸♂️🤸♂️.
Check out the new paper: www.cell.com/molecular-ce...

@roisnehamelinf.bsky.social & co discovered that Wadjet, an SMC complex involved in bacterial DNA immunity, performs some impressive molecular gymnastics 🤸♂️🤸♂️🤸♂️.
Check out the new paper: www.cell.com/molecular-ce...
Deadline for abstracts Oct 15
Final deadline Oct 31
cryoem-symposium.pages.ist.ac.at
Deadline for abstracts Oct 15
Final deadline Oct 31
cryoem-symposium.pages.ist.ac.at
t.co/962Kj9pt6F
t.co/962Kj9pt6F
Top speakers, cutting-edge cryo-EM, and a chance to explore Vienna & the ISTA campus.
Register now 👉 cryoem-symposium.pages.ist.ac.at #cryoEM #teamtomo

Top speakers, cutting-edge cryo-EM, and a chance to explore Vienna & the ISTA campus.
Register now 👉 cryoem-symposium.pages.ist.ac.at #cryoEM #teamtomo
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
We found that by using ab initio reconstruction at very high res, in very small steps, we could crack some small structures that had eluded us - e.g. 39kDa iPKAc (EMPIAR-10252), below.
Read on for details... 1/x
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
Here, we present a method for designing protein nanoparticles with two unique faces that can bring together distinct biological targets at controlled distances!
This project was led by Dr. Sanela Rankovic is out now in Nature Materials: www.nature.com/articles/s41...
Here, we present a method for designing protein nanoparticles with two unique faces that can bring together distinct biological targets at controlled distances!
This project was led by Dr. Sanela Rankovic is out now in Nature Materials: www.nature.com/articles/s41...
We noticed a pair of genes - a nuclease and a protease - shuffles between antiviral systems. We show how proteolysis activates the nuclease, triggering defense in known and unknown immune contexts.
tinyurl.com/2uwwy4ty

We noticed a pair of genes - a nuclease and a protease - shuffles between antiviral systems. We show how proteolysis activates the nuclease, triggering defense in known and unknown immune contexts.
tinyurl.com/2uwwy4ty
We report in @science.org the discovery of a human homolog of SIR2 antiphage proteins that participates in the TLR pathway of animal innate immunity.
Co-led wt @enzopoirier.bsky.social by D. Bonhomme and @hugovaysset.bsky.social
www.science.org/doi/10.1126/...
We report in @science.org the discovery of a human homolog of SIR2 antiphage proteins that participates in the TLR pathway of animal innate immunity.
Co-led wt @enzopoirier.bsky.social by D. Bonhomme and @hugovaysset.bsky.social
www.science.org/doi/10.1126/...
www.nature.com/articles/s41...
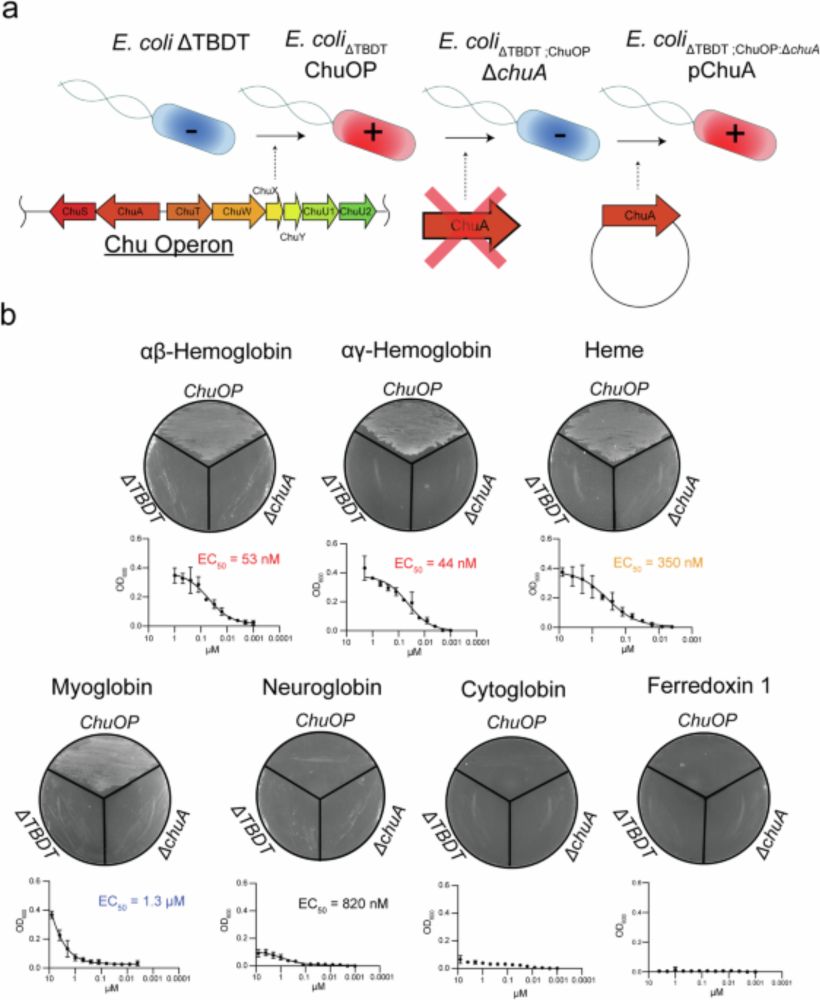
www.nature.com/articles/s41...
🎉Super excited about this milestone as PhD, it’s been a journey, and I’m grateful to finally have these as my first Cryo-EM structures! 🙌
🔁Thank you for sharing!❤️
Read @daaninthelab.bsky.social et. al.: www.science.org/doi/10.1126/...
⬇️ 2nd video
🎉Super excited about this milestone as PhD, it’s been a journey, and I’m grateful to finally have these as my first Cryo-EM structures! 🙌
🔁Thank you for sharing!❤️
Read @daaninthelab.bsky.social et. al.: www.science.org/doi/10.1126/...
⬇️ 2nd video
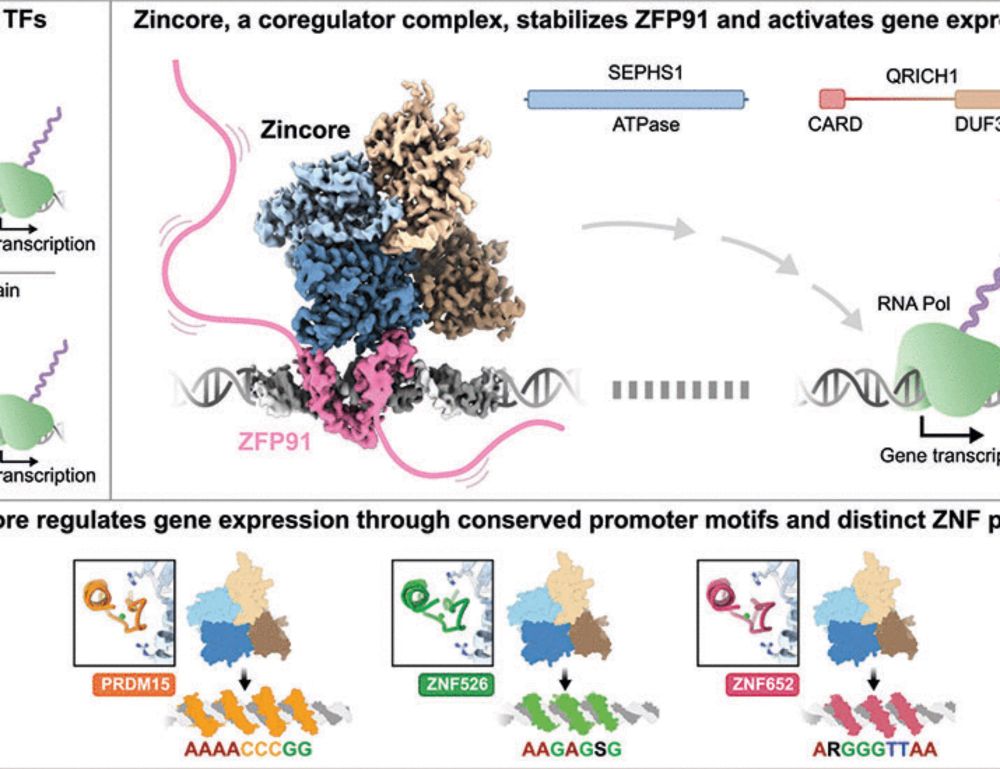
www.nature.com/articles/s41...
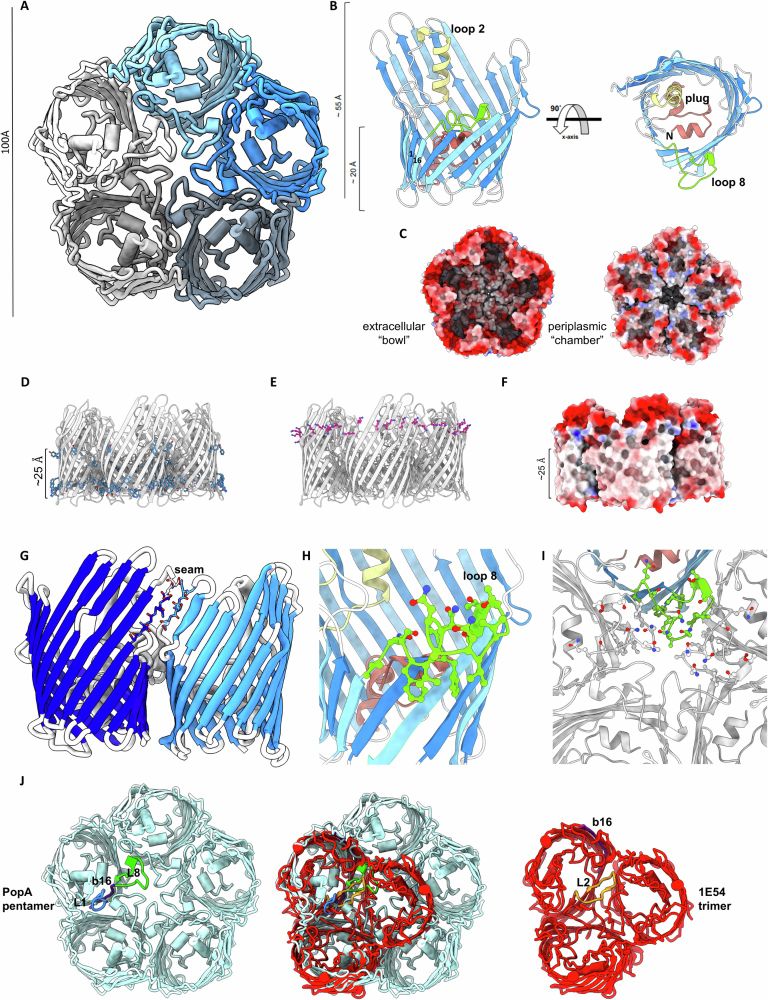
www.nature.com/articles/s41...
Join our ActinID project to explore an uncharacterized actin-binding protein.
- Background in cell and/or structural biology?
- Eager to bridge both fields?
Get in touch if you're curious or have questions!
#cellbiology #cryoEM #cryoET #actin
ista.ac.at/en/job/postd...

Join our ActinID project to explore an uncharacterized actin-binding protein.
- Background in cell and/or structural biology?
- Eager to bridge both fields?
Get in touch if you're curious or have questions!
#cellbiology #cryoEM #cryoET #actin
ista.ac.at/en/job/postd...

