Oli Clarke
@olibclarke.bsky.social
CryoEM, membrane proteins and whatnot
Reposted by Oli Clarke
Announcing cryo-EM heterogeneity challenge #2, now dubbed the 2025 Community-Wide Assessment of Cryo-EM Heterogeneous Reconstruction Algorithms (CAHRA)! Join us for a webinar next Friday (Nov 14th) to learn more. Datasets already posted here: heterogeneity.notion.site/challenge #cryoem

CAHRA 2025: Community-Wide Assessment of Cryo-EM Heterogeneous Reconstruction Algorithms
A community-wide data processing challenge for cryo-electron microscopy.
heterogeneity.notion.site
November 7, 2025 at 10:45 PM
Announcing cryo-EM heterogeneity challenge #2, now dubbed the 2025 Community-Wide Assessment of Cryo-EM Heterogeneous Reconstruction Algorithms (CAHRA)! Join us for a webinar next Friday (Nov 14th) to learn more. Datasets already posted here: heterogeneity.notion.site/challenge #cryoem
Reposted by Oli Clarke
Our November issue is available at journals.iucr.org/d/issues/202...
On the cover: recent studies demonstrate that the range of samples suitable for cryo-EM single-particle analysis is expanding towards increasingly more native samples. Read the review at shorturl.at/mitjg
On the cover: recent studies demonstrate that the range of samples suitable for cryo-EM single-particle analysis is expanding towards increasingly more native samples. Read the review at shorturl.at/mitjg

November 5, 2025 at 1:29 PM
Our November issue is available at journals.iucr.org/d/issues/202...
On the cover: recent studies demonstrate that the range of samples suitable for cryo-EM single-particle analysis is expanding towards increasingly more native samples. Read the review at shorturl.at/mitjg
On the cover: recent studies demonstrate that the range of samples suitable for cryo-EM single-particle analysis is expanding towards increasingly more native samples. Read the review at shorturl.at/mitjg
Reposted by Oli Clarke
We are looking for a computational postdoc to work with us on new optimisation algorithms to make #RELION even better. Join our bubbly team at the @mrclmb.bsky.social in Cambridge, UK. 🤗 RTs appreciated.
mrc.tal.net/vx/appcentre...
mrc.tal.net/vx/appcentre...
Quick Check Needed
mrc.tal.net
October 29, 2025 at 9:17 AM
We are looking for a computational postdoc to work with us on new optimisation algorithms to make #RELION even better. Join our bubbly team at the @mrclmb.bsky.social in Cambridge, UK. 🤗 RTs appreciated.
mrc.tal.net/vx/appcentre...
mrc.tal.net/vx/appcentre...
Reposted by Oli Clarke
New online: New York City’s structural biology mosaic
New York City’s structural biology mosaic
Nature Structural & Molecular Biology, Published online: 28 October 2025; doi:10.1038/s41594-025-01698-zThe structural biology community in New York City combines expertise and access to cutting-edge instrumentation that fosters cooperation. Working collaboratively is indispensable because developing interdisciplinary tools can enable discoveries in cell, structural and molecular biology.
go.nature.com
October 28, 2025 at 11:45 AM
New online: New York City’s structural biology mosaic
Reposted by Oli Clarke
OpenFold3-preview (OF3p) is out: a sneak peek of our AF3-based structure prediction model. Our aim for OF3 is full AF3-parity for every modality. We now believe we have a clear path towards this goal and are releasing OF3p to enable building in the OF3 ecosystem. More👇
October 28, 2025 at 6:30 PM
OpenFold3-preview (OF3p) is out: a sneak peek of our AF3-based structure prediction model. Our aim for OF3 is full AF3-parity for every modality. We now believe we have a clear path towards this goal and are releasing OF3p to enable building in the OF3 ecosystem. More👇
Reposted by Oli Clarke
Lab’s first paper is out!! We show the first structures of #Asgard #chromatin by #cryo-EM 🧬❄️
Asgard histones form closed and open hypernucleosomes. Closed are conserved across #Archaea, while open resemble eukaryotic H3–H4 octasomes and are Asgard-specific. More here: www.cell.com/molecular-ce...
Asgard histones form closed and open hypernucleosomes. Closed are conserved across #Archaea, while open resemble eukaryotic H3–H4 octasomes and are Asgard-specific. More here: www.cell.com/molecular-ce...

October 28, 2025 at 3:07 PM
Lab’s first paper is out!! We show the first structures of #Asgard #chromatin by #cryo-EM 🧬❄️
Asgard histones form closed and open hypernucleosomes. Closed are conserved across #Archaea, while open resemble eukaryotic H3–H4 octasomes and are Asgard-specific. More here: www.cell.com/molecular-ce...
Asgard histones form closed and open hypernucleosomes. Closed are conserved across #Archaea, while open resemble eukaryotic H3–H4 octasomes and are Asgard-specific. More here: www.cell.com/molecular-ce...
Reposted by Oli Clarke
Inhibition mechanism of pancreatic KATP channels by centipede toxins www.biorxiv.org/content/10.1101/2025.10.27.684718v1 #cryoem
October 28, 2025 at 9:28 AM
Inhibition mechanism of pancreatic KATP channels by centipede toxins www.biorxiv.org/content/10.1101/2025.10.27.684718v1 #cryoem
Reposted by Oli Clarke
A class like no other!! From #AI structure hallucination 🤖 to #CryoEM structure reality 🔬 by @biozentrum.unibas.ch @unibas.ch undergraduate students👩🔬👩🎓 in just a few weeks!
One of their creations is this beautiful flower-shaped tetrameric pore 🌼, a brand new member of the #ProteinCosmos 🧶🧬 🧪
One of their creations is this beautiful flower-shaped tetrameric pore 🌼, a brand new member of the #ProteinCosmos 🧶🧬 🧪

October 26, 2025 at 1:48 PM
A class like no other!! From #AI structure hallucination 🤖 to #CryoEM structure reality 🔬 by @biozentrum.unibas.ch @unibas.ch undergraduate students👩🔬👩🎓 in just a few weeks!
One of their creations is this beautiful flower-shaped tetrameric pore 🌼, a brand new member of the #ProteinCosmos 🧶🧬 🧪
One of their creations is this beautiful flower-shaped tetrameric pore 🌼, a brand new member of the #ProteinCosmos 🧶🧬 🧪
Reposted by Oli Clarke
October release of DAQ Score DB of cryo-EM str validation! Now includes 255,199 protein chains from 17,283 EMDB maps. Fig is an example with an entire helix having a residue shift.
daqdb.kiharalab.org
Easy to use DAQ for structure validation in your paper: em.kiharalab.org
daqdb.kiharalab.org
Easy to use DAQ for structure validation in your paper: em.kiharalab.org

October 26, 2025 at 7:23 PM
October release of DAQ Score DB of cryo-EM str validation! Now includes 255,199 protein chains from 17,283 EMDB maps. Fig is an example with an entire helix having a residue shift.
daqdb.kiharalab.org
Easy to use DAQ for structure validation in your paper: em.kiharalab.org
daqdb.kiharalab.org
Easy to use DAQ for structure validation in your paper: em.kiharalab.org
Reposted by Oli Clarke
Excited to release BoltzGen which brings SOTA folding performance to binder design! The best part of this project is collaborating with a broad network of leading wetlabs that test BoltzGen at an unprecedented scale, showing success on many novel targets and pushing the model to its limits!

October 26, 2025 at 10:40 PM
Excited to release BoltzGen which brings SOTA folding performance to binder design! The best part of this project is collaborating with a broad network of leading wetlabs that test BoltzGen at an unprecedented scale, showing success on many novel targets and pushing the model to its limits!
Reposted by Oli Clarke
I love AlphaFold—but please include PAE (Predicted Aligned Error) plots for every “interaction.” Pretty PDBs ≠ proof. If the PAE doesn’t show an interface, it ain’t one. Show the plots. Structural biology's reputation is on the line.
October 24, 2025 at 5:44 PM
I love AlphaFold—but please include PAE (Predicted Aligned Error) plots for every “interaction.” Pretty PDBs ≠ proof. If the PAE doesn’t show an interface, it ain’t one. Show the plots. Structural biology's reputation is on the line.
Reposted by Oli Clarke
We are excited to share our latest work on GLIC published in @pnas.org, where we developed a #cryoEM data processing strategy that allowed us to isolate and reconstruct multiple asymmetric states, revealing detailed insights into the channel’s activation mechanism. tinyurl.com/mwtb4rpj

Asymmetric gating of a homopentameric ion channel GLIC revealed by cryo-EM | PNAS
Pentameric ligand-gated ion channels (pLGICs) are vital neurotransmitter receptors
that are key therapeutic targets for neurological disorders. Alt...
tinyurl.com
October 24, 2025 at 5:32 AM
We are excited to share our latest work on GLIC published in @pnas.org, where we developed a #cryoEM data processing strategy that allowed us to isolate and reconstruct multiple asymmetric states, revealing detailed insights into the channel’s activation mechanism. tinyurl.com/mwtb4rpj
Reposted by Oli Clarke
Helicon: Helical parameter determination and 3D reconstruction from one image pubmed.ncbi.nlm.nih.gov/41130582/ #cryoem
October 24, 2025 at 2:50 PM
Helicon: Helical parameter determination and 3D reconstruction from one image pubmed.ncbi.nlm.nih.gov/41130582/ #cryoem
Reposted by Oli Clarke
Finally out in print! Long time project with @olexandr.bsky.social and the PHENIX group. Refinement of X-ray and CryoEM structures, using Machine Learned Potentials as the back end to help fitting. This fulfills the old dream of doing QM refinement, but very very cheap www.nature.com/articles/s41...

AQuaRef: machine learning accelerated quantum refinement of protein structures - Nature Communications
AQuaRef employs machine learning to refine protein structures from cryo-EM and X-ray data in Phenix. It achieves quantum-level precision, improving model geometry and fit to the data while reducing ov...
www.nature.com
October 23, 2025 at 3:20 PM
Finally out in print! Long time project with @olexandr.bsky.social and the PHENIX group. Refinement of X-ray and CryoEM structures, using Machine Learned Potentials as the back end to help fitting. This fulfills the old dream of doing QM refinement, but very very cheap www.nature.com/articles/s41...
Reposted by Oli Clarke
Visualizing influenza A virus assembly by in situ cryo-electron tomography
www.nature.com/articles/s41...
www.nature.com/articles/s41...
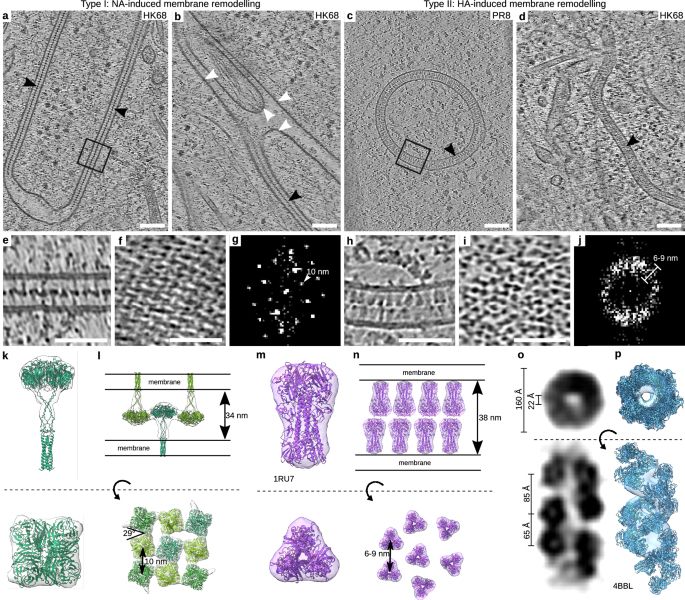
Visualizing influenza A virus assembly by in situ cryo-electron tomography - Nature Communications
Influenza A virus must package eight separate genomic segments, called viral ribonucleoproteins (vRNPs). Using in situ cryo-electron tomography, the authors visualize how vRNPs are clustered on cell m...
www.nature.com
October 23, 2025 at 2:01 PM
Visualizing influenza A virus assembly by in situ cryo-electron tomography
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Reposted by Oli Clarke
“Kiss-shrink-run” unifies mechanisms for synaptic vesicle exocytosis and hyperfast recycling | Science www.science.org/doi/10.1126/...

“Kiss-shrink-run” unifies mechanisms for synaptic vesicle exocytosis and hyperfast recycling
Synaptic vesicle (SV) exocytosis underpins neuronal communication, yet its nanoscale dynamics remain poorly understood owing to limitations in visualizing rapid events in situ. Here, we used optogenet...
www.science.org
October 17, 2025 at 12:54 PM
“Kiss-shrink-run” unifies mechanisms for synaptic vesicle exocytosis and hyperfast recycling | Science www.science.org/doi/10.1126/...
Wild
A biased allosteric modulator functions as a molecular glue to induce β2AR dimerization www.biorxiv.org/content/10.1101/2025.10.21.683802v1 #cryoem
October 23, 2025 at 10:40 AM
Wild
Reposted by Oli Clarke
Chris Russo and Anastasiia Gusach have developed a new method to prepare #cryoEM samples that avoids protein damage during freezing
Read more: www2.mrc-lmb.cam.ac.uk/a-new-method...
#LMBResearch
Read more: www2.mrc-lmb.cam.ac.uk/a-new-method...
#LMBResearch
October 22, 2025 at 4:41 PM
Chris Russo and Anastasiia Gusach have developed a new method to prepare #cryoEM samples that avoids protein damage during freezing
Read more: www2.mrc-lmb.cam.ac.uk/a-new-method...
#LMBResearch
Read more: www2.mrc-lmb.cam.ac.uk/a-new-method...
#LMBResearch
Reposted by Oli Clarke
End-to-end protein design in the browser through evedesign. Generate and interactively explore designs in 2D/3D and export them as codon-optimized DNA. The underlying open source framework (released soon) is build to easily add new methods, more on that soon.
🌐 evedesign.bio
🌐 evedesign.bio
October 22, 2025 at 2:30 PM
End-to-end protein design in the browser through evedesign. Generate and interactively explore designs in 2D/3D and export them as codon-optimized DNA. The underlying open source framework (released soon) is build to easily add new methods, more on that soon.
🌐 evedesign.bio
🌐 evedesign.bio
Reposted by Oli Clarke
**Job Alert** Exciting postdoc position available: theoretical and experimental cryo-EM studies of flexible biomolecules. Competitive salary, collaborative environment at NYSBC and Flatiron Institute. Please share!! and contact: pcossio@flatironinstitute.org
October 22, 2025 at 2:55 PM
**Job Alert** Exciting postdoc position available: theoretical and experimental cryo-EM studies of flexible biomolecules. Competitive salary, collaborative environment at NYSBC and Flatiron Institute. Please share!! and contact: pcossio@flatironinstitute.org
Is anyone else having problems with NCBI tools? I'm trying to use the CDD database, and it seems completely broken for requests that worked fine before - not sure if shutdown related?

October 21, 2025 at 6:52 PM
Is anyone else having problems with NCBI tools? I'm trying to use the CDD database, and it seems completely broken for requests that worked fine before - not sure if shutdown related?
Reposted by Oli Clarke
Reposted by Oli Clarke
So excited to see this project finally come together after 2+ years of development! I’m proud of my contributions, but this success belongs to the entire Structura team — such a great example of what collaboration and creativity can achieve.
🚀 Introducing fully automated data processing for repeat-target #cryoEM.
Using new tools in #CryoSPARC, it is now possible to obtain resolutions & map quality equal to or better than manual processing, with zero user intervention.
Preprint: www.biorxiv.org/content/10.1...
Using new tools in #CryoSPARC, it is now possible to obtain resolutions & map quality equal to or better than manual processing, with zero user intervention.
Preprint: www.biorxiv.org/content/10.1...
October 20, 2025 at 2:45 PM
So excited to see this project finally come together after 2+ years of development! I’m proud of my contributions, but this success belongs to the entire Structura team — such a great example of what collaboration and creativity can achieve.
Reposted by Oli Clarke
End-to-end automation of repeat-target cryo-EM structure determination in CryoSPARC https://www.biorxiv.org/content/10.1101/2025.10.17.682689v1
October 20, 2025 at 1:47 PM
End-to-end automation of repeat-target cryo-EM structure determination in CryoSPARC https://www.biorxiv.org/content/10.1101/2025.10.17.682689v1
Reposted by Oli Clarke
Learn how our hunt for the native structure of top antimalarial target PfATP4🧂led to the discovery of PfABP, an unknown essential binding partner, out now! @natcomms.nature.com 👉 rdcu.be/eLRlH 🦠🔬❄️
A team effort led by @mehsehret.bsky.social & Anurag Shukla @akhilvaidya.bsky.social! #cryoEM #malaria
A team effort led by @mehsehret.bsky.social & Anurag Shukla @akhilvaidya.bsky.social! #cryoEM #malaria

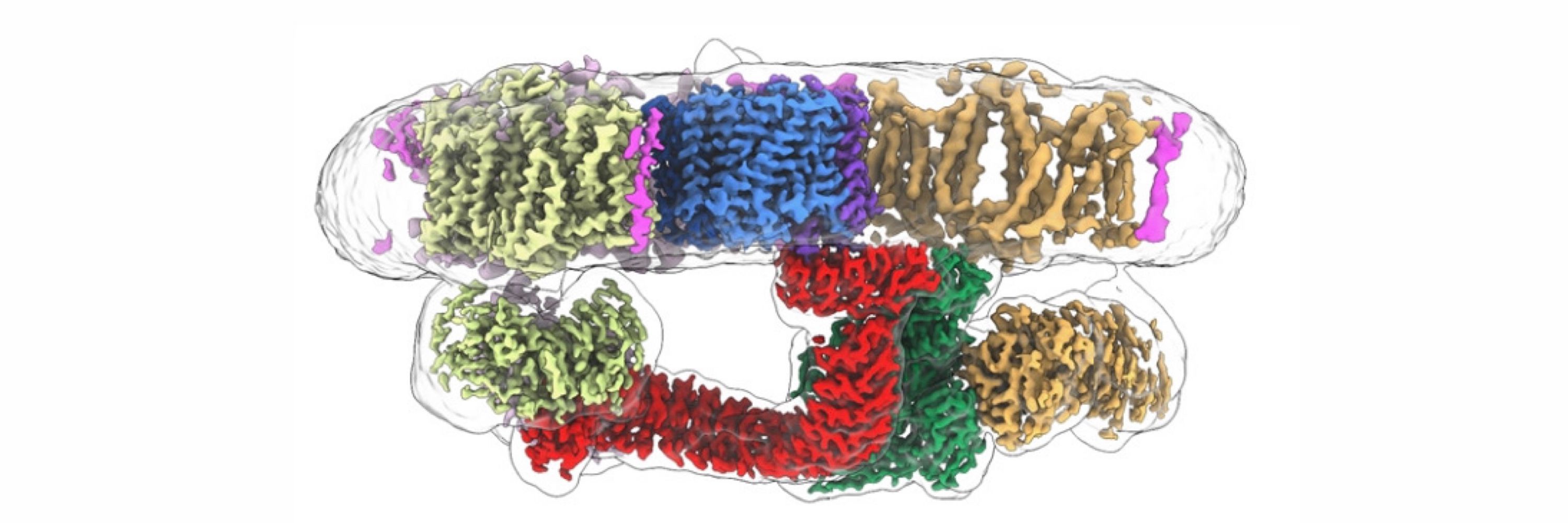
Endogenous structure of antimalarial target PfATP4 reveals an apicomplexan-specific P-type ATPase modulator
Nature Communications - Here, the authors present the 3.7 Å cryoEM structure of native sodium efflux pump PfATP4 from Plasmodium falciparum, revealing a bound protein that they term...
rdcu.be
October 20, 2025 at 12:55 PM
Learn how our hunt for the native structure of top antimalarial target PfATP4🧂led to the discovery of PfABP, an unknown essential binding partner, out now! @natcomms.nature.com 👉 rdcu.be/eLRlH 🦠🔬❄️
A team effort led by @mehsehret.bsky.social & Anurag Shukla @akhilvaidya.bsky.social! #cryoEM #malaria
A team effort led by @mehsehret.bsky.social & Anurag Shukla @akhilvaidya.bsky.social! #cryoEM #malaria


