Assistant Member @MSKCC
Formerly post-doc @MIT
Formerly-formerly PhD @MRC_LMB
Formerly formerly-formerly Otago Uni
Kiwi 🥝 🇳🇿
https://wilkinsonlab.bio/
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Amazing work from Jess Warren (www.hhmi.org/scientists/j...) to figure this out and prove it was real, given the wacky and challenging insect system we've got here.
www.biorxiv.org/content/10.1...

Amazing work from Jess Warren (www.hhmi.org/scientists/j...) to figure this out and prove it was real, given the wacky and challenging insect system we've got here.
www.biorxiv.org/content/10.1...
🧬We'll be finding funky new RNA biology, mainly by looking at reverse transcriptases (i.e. the Best Enzymes In The World)🧬
annnd: I'm hiring - come join! Especially postdocs and PhD students - please get in touch (NYC is great)

🧬We'll be finding funky new RNA biology, mainly by looking at reverse transcriptases (i.e. the Best Enzymes In The World)🧬
annnd: I'm hiring - come join! Especially postdocs and PhD students - please get in touch (NYC is great)
1- other RT phylogenies place TERT within eukaryotic retroelement clades
2- template jumping could plausibly evolve into repeat synthesis activity multiple independent times
Today, we report the discovery of telomerase homologs in a family of antiviral RTs, revealing an unexpected evolutionary origin in bacteria.
www.biorxiv.org/content/10.1...

1- other RT phylogenies place TERT within eukaryotic retroelement clades
2- template jumping could plausibly evolve into repeat synthesis activity multiple independent times
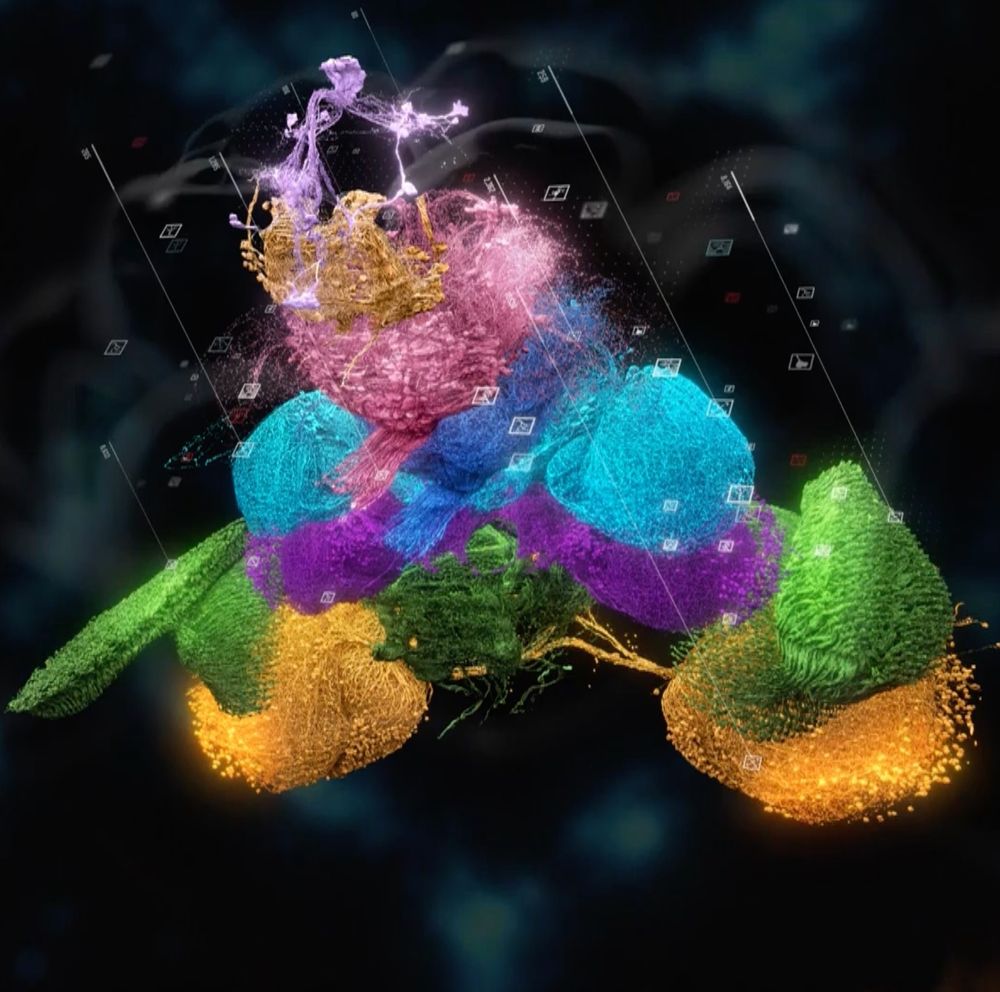
Different from prior connectomes - it is brain + cord (think spinal cord)
We use it to ‘embody’ the system and find it resembles ‘subsumption architecture’ doi.org/10.1101/2025...
Different from prior connectomes - it is brain + cord (think spinal cord)
We use it to ‘embody’ the system and find it resembles ‘subsumption architecture’ doi.org/10.1101/2025...
If you you love genome editing...
Or if you just like random bird animations,
we have the paper for you!
We (@kedmonds.bsky.social et al) are happy to share our work turning a songbird retrotransposon into a genome editing tool. 🐣 (1/n)
If you you love genome editing...
Or if you just like random bird animations,
we have the paper for you!
We (@kedmonds.bsky.social et al) are happy to share our work turning a songbird retrotransposon into a genome editing tool. 🐣 (1/n)

We decided to call them TIGR-Tas systems 🐯
We decided to call them TIGR-Tas systems 🐯
(it's a spliceosome! Truly it was a nostalgic pleasure to help out with this paper, a super fun collaboration with @uwmadisonrna.bsky.social & Kathy and Karli)
https://buff.ly/49aQvWQ

(it's a spliceosome! Truly it was a nostalgic pleasure to help out with this paper, a super fun collaboration with @uwmadisonrna.bsky.social & Kathy and Karli)