https://jacobhepkema.github.io

🔗 biorxiv.org/content/10.1...
A short thread (by Juliane Weller)👇

🔗 biorxiv.org/content/10.1...
A short thread (by Juliane Weller)👇
Are Transcription Factors really 'undruggable'?
www.biorxiv.org/content/10.1...

Are Transcription Factors really 'undruggable'?
www.biorxiv.org/content/10.1...
"De-novo promoters emerge more readily from random DNA than from genomic DNA"
This project is the accumulation of 4 years of work, and lays the foundation for my future group. In short, we… (1/4)

"De-novo promoters emerge more readily from random DNA than from genomic DNA"
This project is the accumulation of 4 years of work, and lays the foundation for my future group. In short, we… (1/4)


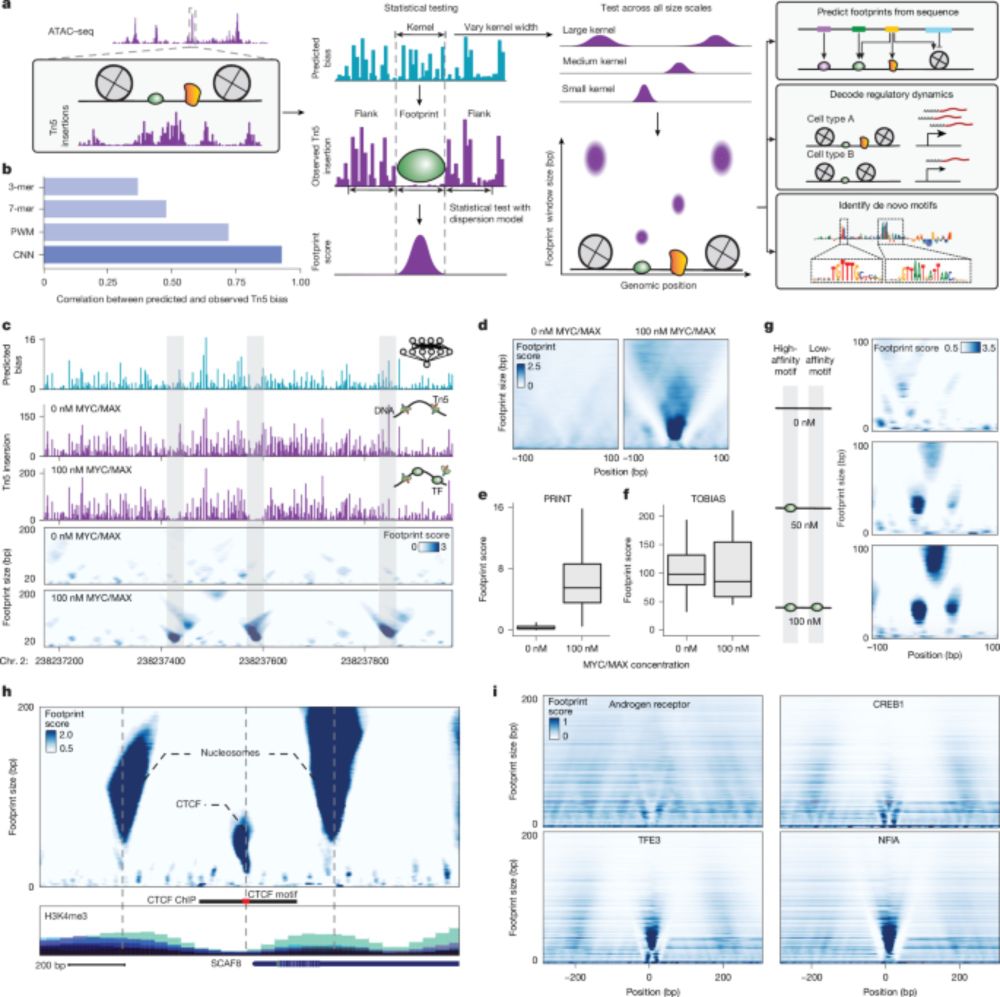
But how do they REALLY work?
New paper with many contributors here @berkeleylab.lbl.gov, @anshulkundaje.bsky.social, @anusri.bsky.social
A 🧵 (1/n)
Free access link: rdcu.be/erD22
But how do they REALLY work?
New paper with many contributors here @berkeleylab.lbl.gov, @anshulkundaje.bsky.social, @anusri.bsky.social
A 🧵 (1/n)
Free access link: rdcu.be/erD22
Check out our paper - fresh off the press!!!
We find widespread functional conservation of enhancers in absence of sequence homology
Including: a bioinformatic tool to map sequence-diverged enhancers!
rdcu.be/enVDN
github.com/tobiaszehnde...

Check out our paper - fresh off the press!!!
We find widespread functional conservation of enhancers in absence of sequence homology
Including: a bioinformatic tool to map sequence-diverged enhancers!
rdcu.be/enVDN
github.com/tobiaszehnde...
Rewriting regulatory DNA to dissect and reprogram gene expression
Our new method (Variant-EFFECTS) uses high-throughput prime editing + flow sorting + sequencing to precisely measure effects of noncoding variants on gene expression
Thread 👇

Rewriting regulatory DNA to dissect and reprogram gene expression
Our new method (Variant-EFFECTS) uses high-throughput prime editing + flow sorting + sequencing to precisely measure effects of noncoding variants on gene expression
Thread 👇
www.nature.com/articles/s41...
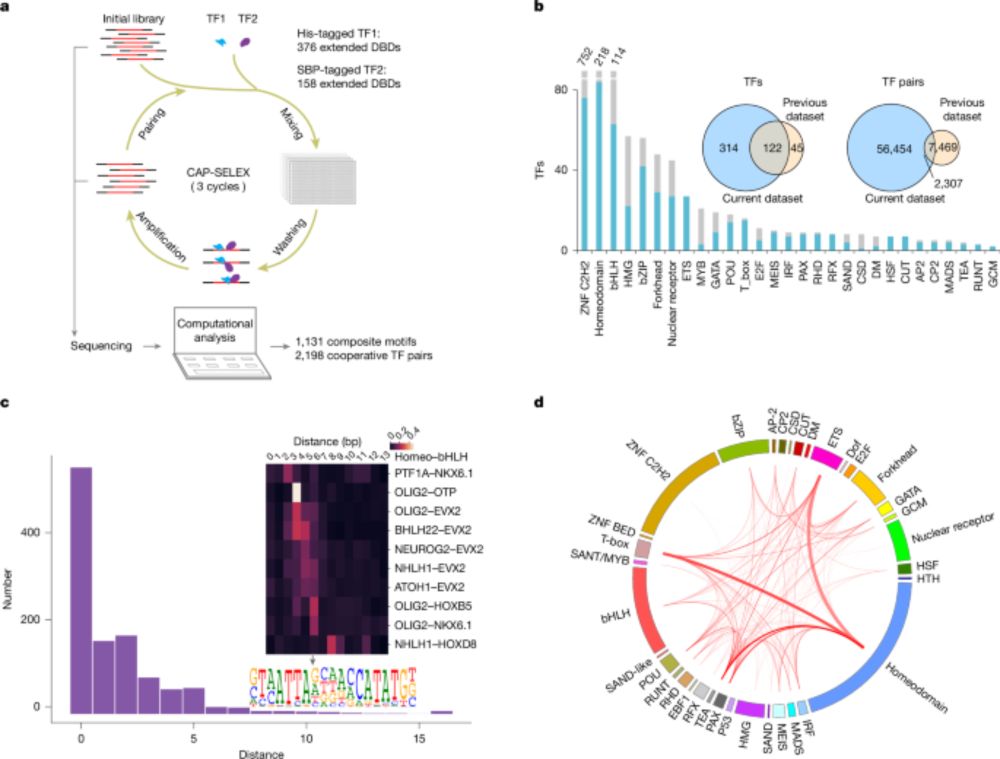
www.nature.com/articles/s41...
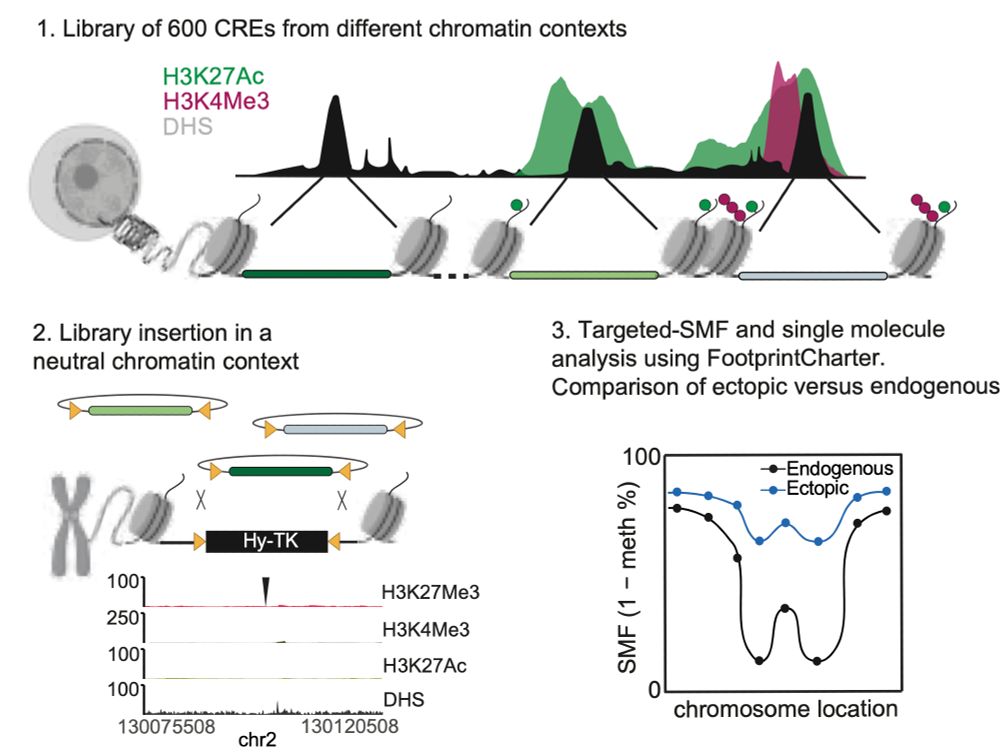
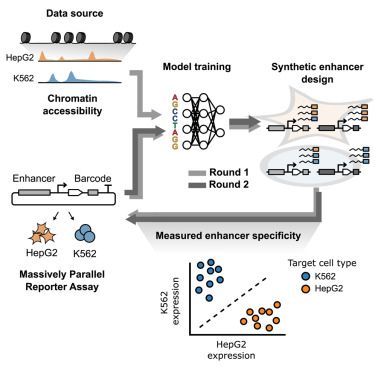
1) CREsted: to train sequence-to-function deep learning models on scATAC-seq atlases, and use them to decipher enhancer logic and design synthetic enhancers. This has been a wonderful lab-wide collaborative effort. www.biorxiv.org/content/10.1...

1) CREsted: to train sequence-to-function deep learning models on scATAC-seq atlases, and use them to decipher enhancer logic and design synthetic enhancers. This has been a wonderful lab-wide collaborative effort. www.biorxiv.org/content/10.1...
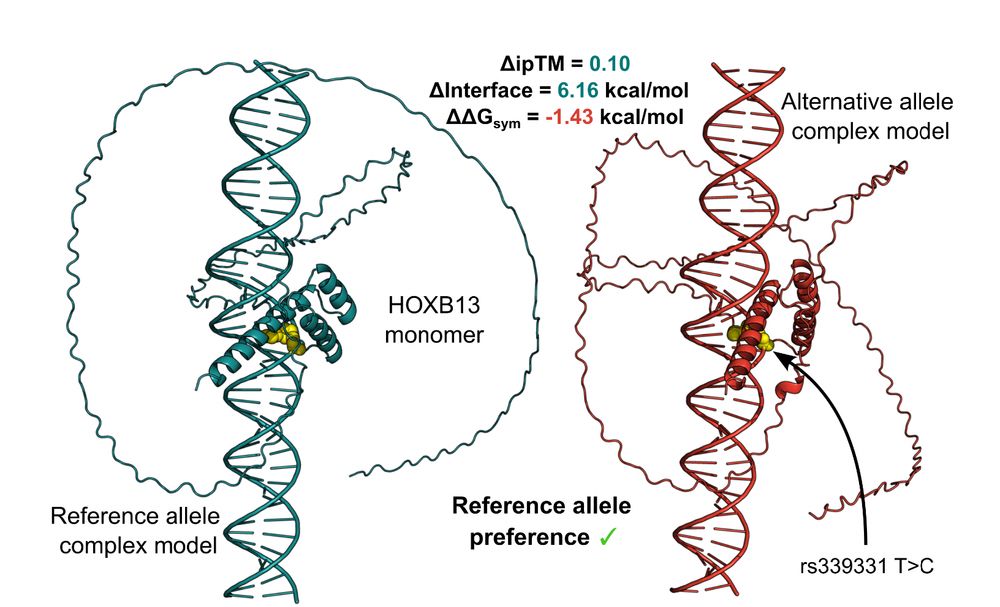
Led by @ioansarr.bsky.social, @marisepp.bsky.social and @tyamadat.bsky.social, in collaboration with @steinaerts.bsky.social
Led by @ioansarr.bsky.social, @marisepp.bsky.social and @tyamadat.bsky.social, in collaboration with @steinaerts.bsky.social
www.science.org/doi/10.1126/...
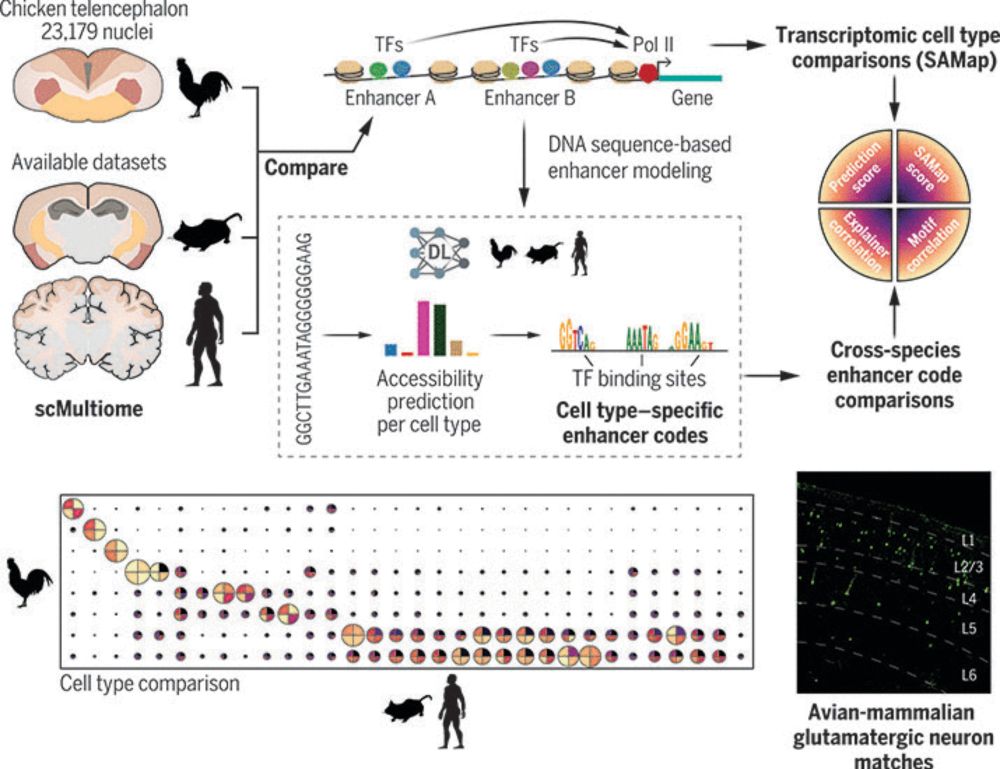
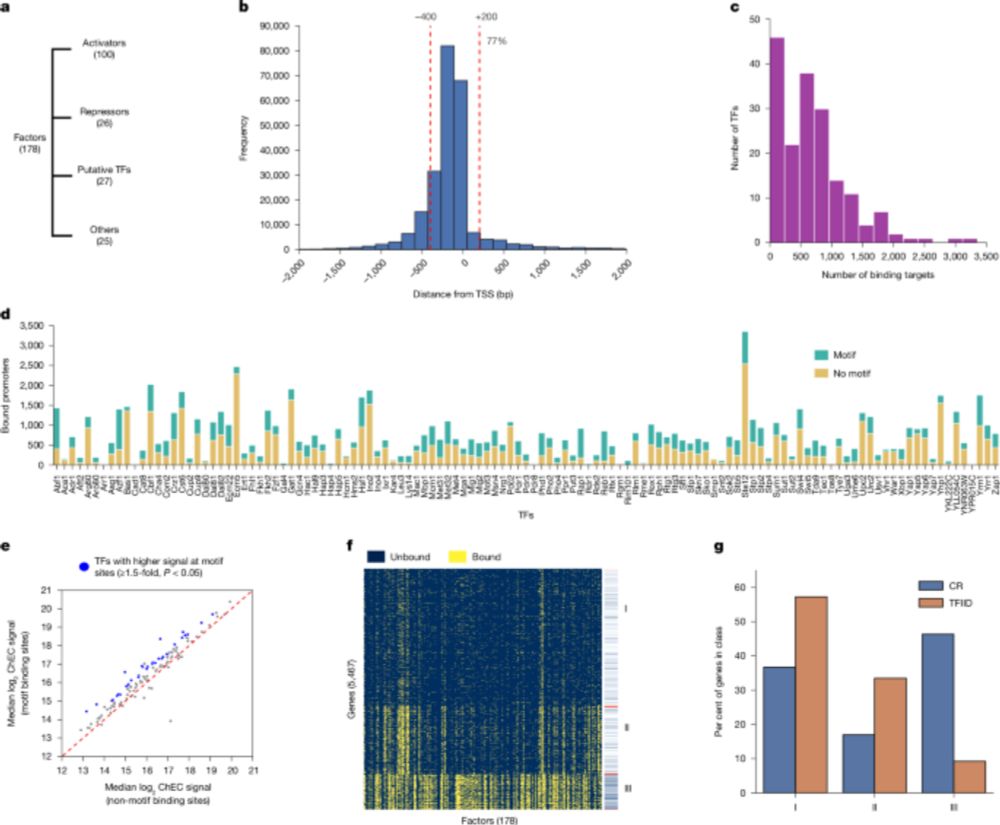
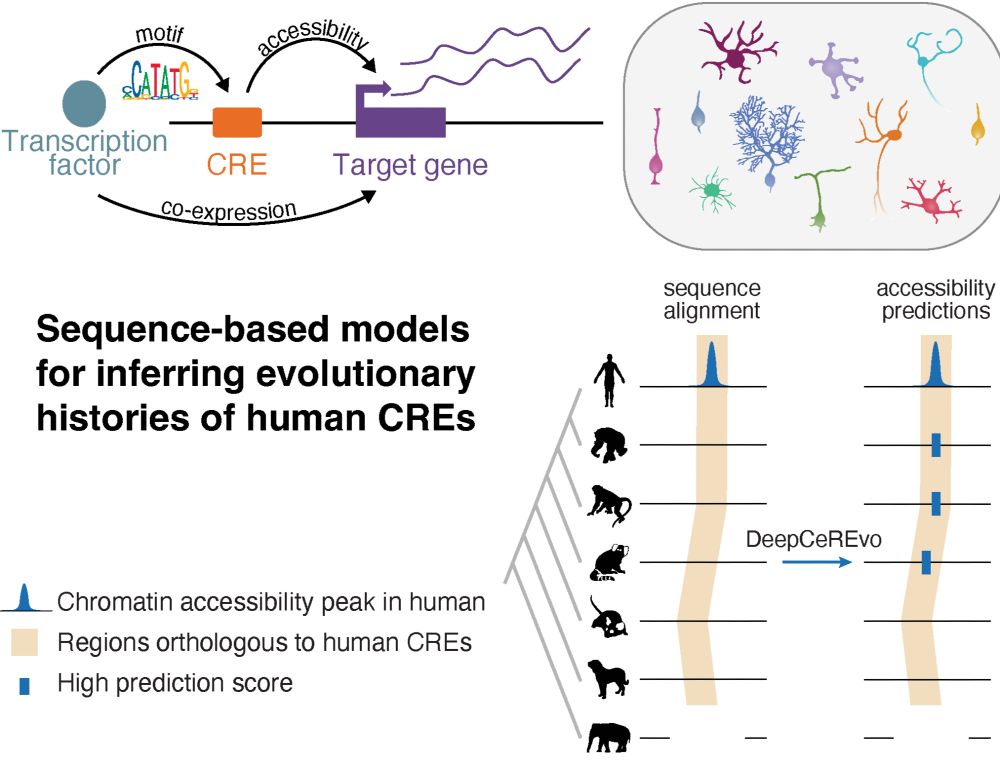
Here are three example catalogs designed for GATA2 binding, chromatin accessibility, and transcription initiation.

Here are three example catalogs designed for GATA2 binding, chromatin accessibility, and transcription initiation.
By investigating bacterial genomes put in yeast, we show that the presence or absence of transcription is sufficient!
#chromatin #3Dgenome #generegulation
www.science.org/doi/10.1126/...
👇
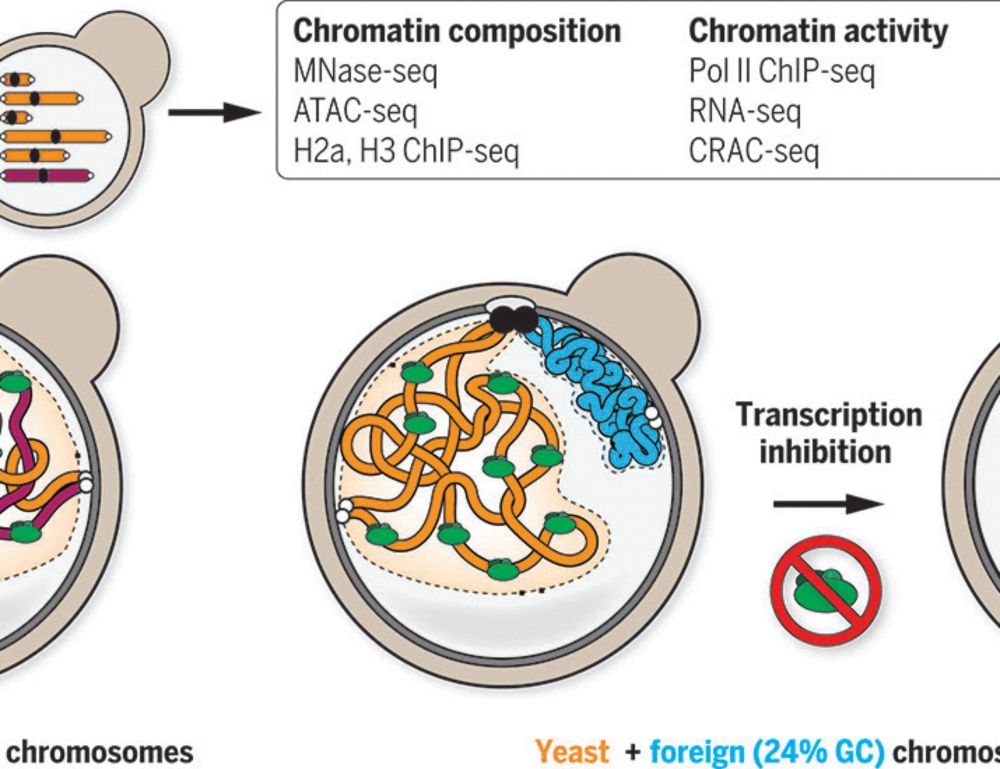
By investigating bacterial genomes put in yeast, we show that the presence or absence of transcription is sufficient!
#chromatin #3Dgenome #generegulation
www.science.org/doi/10.1126/...
👇
rdcu.be/d8xLv (1/6)
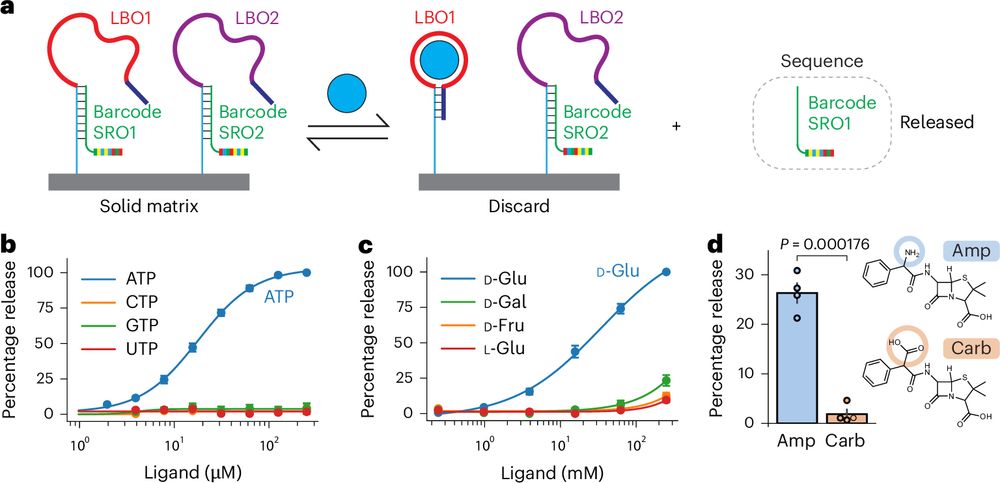
rdcu.be/d8xLv (1/6)
www.science.org/doi/10.1126/...
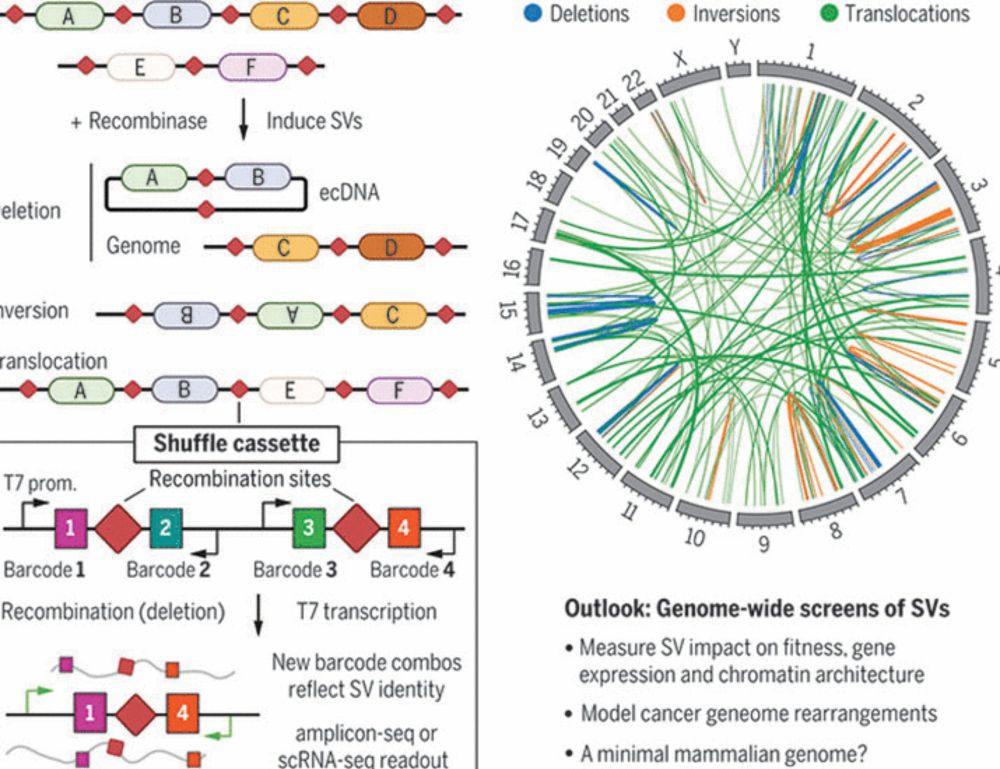
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
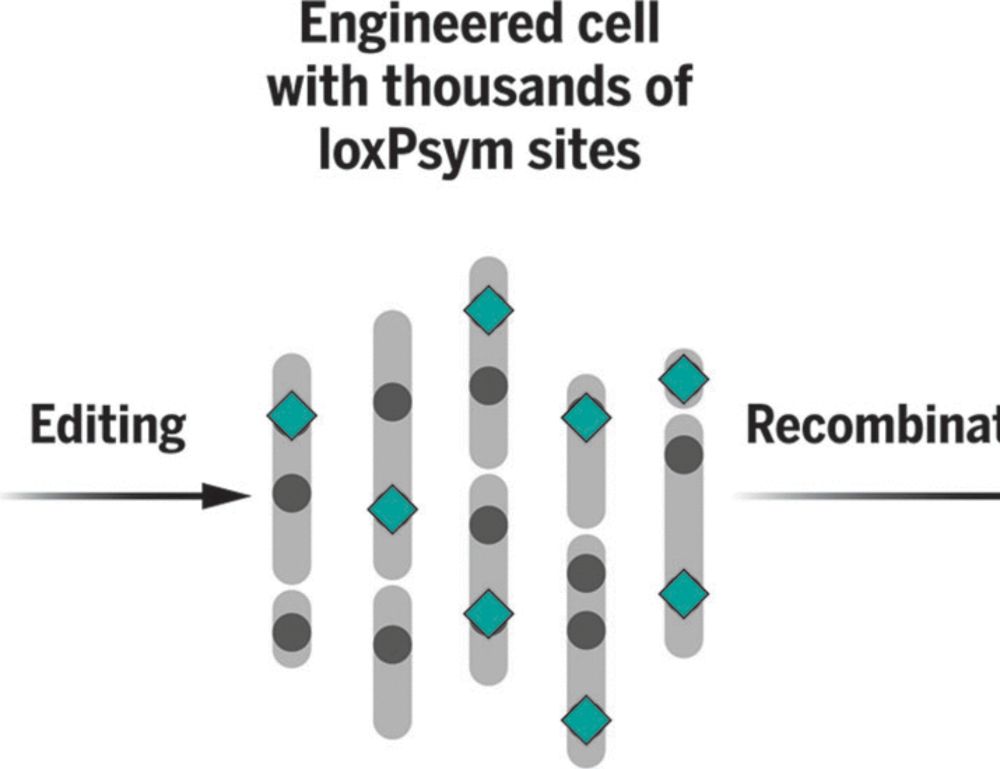
www.science.org/doi/10.1126/...
Paper: www.nature.com/articles/s41...