

Read our paper @cp-cell.bsky.social!
❕Publication: doi.org/10.1016/j.ce...
❕Press Release: www.biochem.mpg.de/en/pressroom
@uoftmedicine.bsky.social
@erc.europa.eu #UPSmeetMet
Read our paper @cp-cell.bsky.social!
❕Publication: doi.org/10.1016/j.ce...
❕Press Release: www.biochem.mpg.de/en/pressroom
@uoftmedicine.bsky.social
@erc.europa.eu #UPSmeetMet


Super cool mechanism of how a metabolite regulates the stability of its own metabolizing enzyme!
Alina did it all for this project CRISPR screen ✂️, biochemistry 🧪, and cryo-EM ❄️🔬.
Congrats!
We are excited to share our recent work on #E3 ligase regulation in #metabolism!
www.biorxiv.org/content/10.6...
#ubiquitin #targetedproteindegradation #chemicalbiology
1/6
Super cool mechanism of how a metabolite regulates the stability of its own metabolizing enzyme!
Alina did it all for this project CRISPR screen ✂️, biochemistry 🧪, and cryo-EM ❄️🔬.
Congrats!
Massive congrats to this transatlantic collaboration between @jakobfarnung.bsky.social from Schulman Lab and @elenaslo.bsky.social from @bartellab.bsky.social
Have a read!
Massive congrats to this transatlantic collaboration between @jakobfarnung.bsky.social from Schulman Lab and @elenaslo.bsky.social from @bartellab.bsky.social
Have a read!
Massive Congrats @jakobfarnung.bsky.social and collaborators from @bartellab.bsky.social
Massive Congrats @jakobfarnung.bsky.social and collaborators from @bartellab.bsky.social


Many thanks to @lisaheinke.bsky.social for the opportunity to write this TotT!

Many thanks to @lisaheinke.bsky.social for the opportunity to write this TotT!

1/9
www.nature.com/articles/s41...
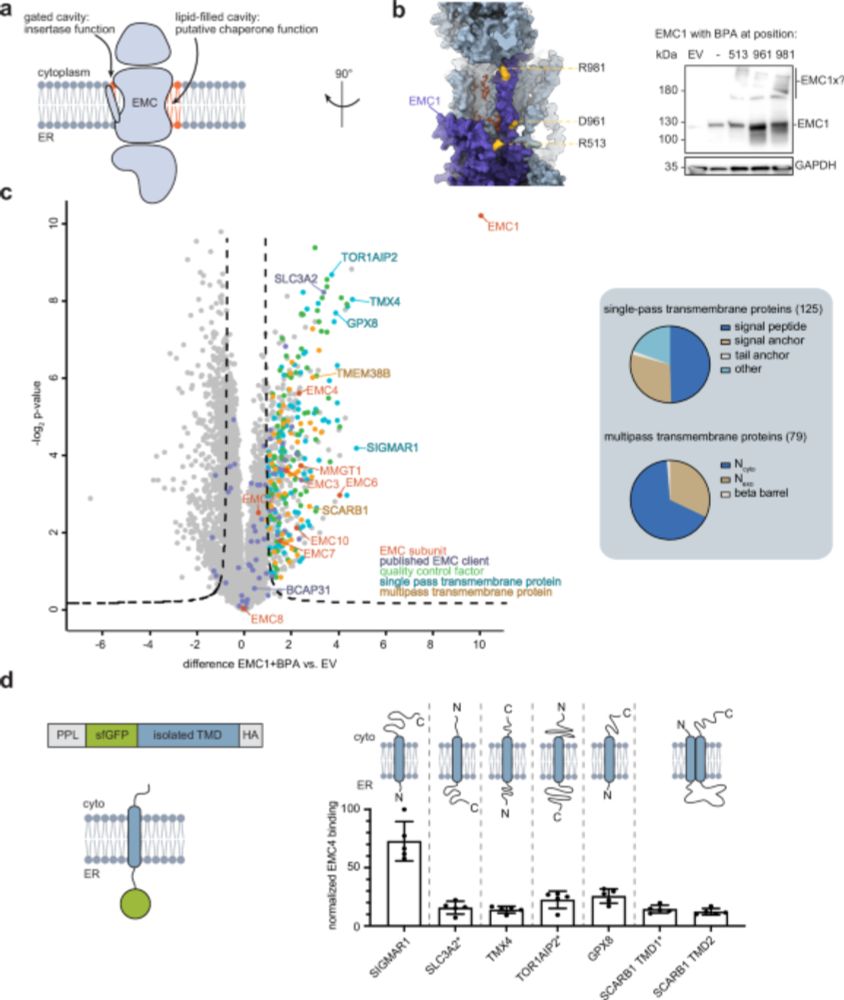
1/9
www.nature.com/articles/s41...
Then join us tomorrow for the latest instalment of the @danafarber.bsky.social Targeted Protein Degradation Webinar where Kimberly Stegmaier and I will present our latest work.
dfci.zoom.us/webinar/regi...

Then join us tomorrow for the latest instalment of the @danafarber.bsky.social Targeted Protein Degradation Webinar where Kimberly Stegmaier and I will present our latest work.
dfci.zoom.us/webinar/regi...

#wUeBI2025 @grk2243.bsky.social
@jakobfarnung.bsky.social @samuelmaiwald.bsky.social @hannahbkmpr.bsky.social

#wUeBI2025 @grk2243.bsky.social
@jakobfarnung.bsky.social @samuelmaiwald.bsky.social @hannahbkmpr.bsky.social
@natsmb.nature.com
1/7
www.nature.com/articles/s41...
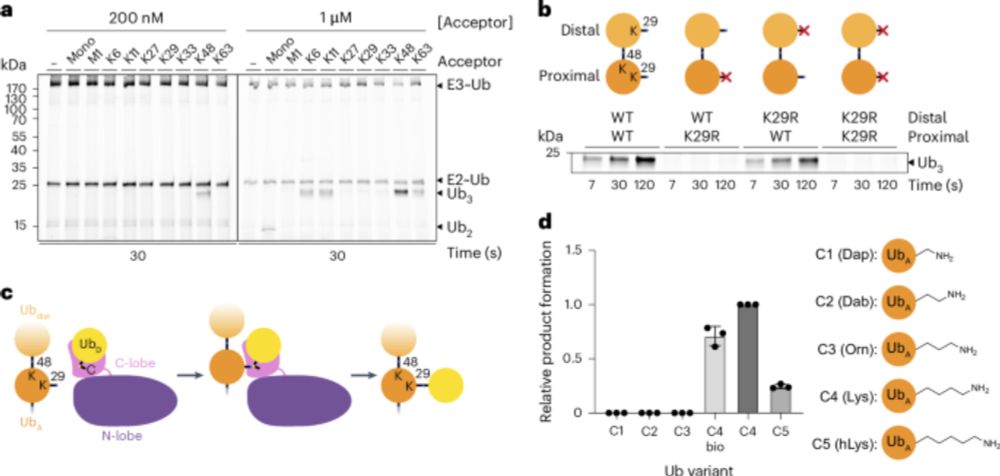
@natsmb.nature.com
1/7
www.nature.com/articles/s41...

@natsmb.nature.com
1/7
www.nature.com/articles/s41...

