
Scientist at @mpibiochem.bsky.social
Alumnus of @mrclmb.bsky.social
Have a look at my recent talk at the @danafarber.bsky.social Targeted Protein Degradation Webinar Series now out on YouTube. Here, I talk about my recent publication (see Blue-torial below)
www.youtube.com/watch?v=Cpg0...



Read our paper @cp-cell.bsky.social!
❕Publication: doi.org/10.1016/j.ce...
❕Press Release: www.biochem.mpg.de/en/pressroom
@uoftmedicine.bsky.social
@erc.europa.eu #UPSmeetMet
Read our paper @cp-cell.bsky.social!
❕Publication: doi.org/10.1016/j.ce...
❕Press Release: www.biochem.mpg.de/en/pressroom
@uoftmedicine.bsky.social
@erc.europa.eu #UPSmeetMet
Using cryo-electron tomography, we show it forms amyloid structures inside lysosomes that mechanically rupture membranes – revealing a new paradigm for lysosomal failure.
🔗 doi.org/10.64898/202...
#CryoET

Using cryo-electron tomography, we show it forms amyloid structures inside lysosomes that mechanically rupture membranes – revealing a new paradigm for lysosomal failure.
🔗 doi.org/10.64898/202...
#CryoET
We took advantage of the MitoTag mouse line to study what mRNAs associate with mitochondria in axons in vivo, and we were in for a surprise! Other than our favorite Pink1, mainly cytoskeletal mRNAs hitch a ride and pave the way for axon growth. shorturl.at/rYe1U

We took advantage of the MitoTag mouse line to study what mRNAs associate with mitochondria in axons in vivo, and we were in for a surprise! Other than our favorite Pink1, mainly cytoskeletal mRNAs hitch a ride and pave the way for axon growth. shorturl.at/rYe1U
We are excited to share our recent work on #E3 ligase regulation in #metabolism!
www.biorxiv.org/content/10.6...
#ubiquitin #targetedproteindegradation #chemicalbiology
1/6
Fantastic guest speakers👇 & many slots for talks from abstracts, flash-talks & posters. Lots of opportunities to network. Register now to save your spot!
➡️ www.protein-degradation.org/symposium/
#ubfriends2026

Massive congrats to this transatlantic collaboration between @jakobfarnung.bsky.social from Schulman Lab and @elenaslo.bsky.social from @bartellab.bsky.social
Have a read!
Massive congrats to this transatlantic collaboration between @jakobfarnung.bsky.social from Schulman Lab and @elenaslo.bsky.social from @bartellab.bsky.social
Have a read!
Massive Congrats @jakobfarnung.bsky.social and collaborators from @bartellab.bsky.social
Massive Congrats @jakobfarnung.bsky.social and collaborators from @bartellab.bsky.social


We have developed a new strategy termed "potency coherence analysis" that leverages the drug potency dimension in decryptM to decode the kinases that shape the human phosphoproteome.
Read more:

We have developed a new strategy termed "potency coherence analysis" that leverages the drug potency dimension in decryptM to decode the kinases that shape the human phosphoproteome.
Read more:


A project on protein binder discovery for K6-linked #ubiquitin to investigate their role in infection is available.
📍 DTU, Copenhagen, Denmark
⏰ Deadline: 18 December 2025
Apply here: efzu.fa.em2.oraclecloud.com/hcmUI/Candid...

A project on protein binder discovery for K6-linked #ubiquitin to investigate their role in infection is available.
📍 DTU, Copenhagen, Denmark
⏰ Deadline: 18 December 2025
Apply here: efzu.fa.em2.oraclecloud.com/hcmUI/Candid...
Our new paper investigates the nuclear events leading to human mRNA export. www.nature.com/articles/s41.... (1/4)
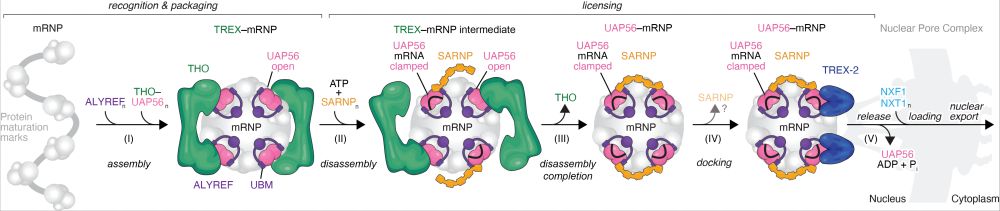
Our new paper investigates the nuclear events leading to human mRNA export. www.nature.com/articles/s41.... (1/4)
Many thanks to @lisaheinke.bsky.social for the opportunity to write this TotT!

