
Koina aims to transform how #proteomics uses machine learning. You no longer need to be a tech wizard to use ML and now can easily run #ML models. Integrated with FragPipe, Skyline and EncyclopeDIA!
www.nature.com/articles/s41...

Koina aims to transform how #proteomics uses machine learning. You no longer need to be a tech wizard to use ML and now can easily run #ML models. Integrated with FragPipe, Skyline and EncyclopeDIA!
www.nature.com/articles/s41...
Data-augmented DL enables zero-shot PTM prediction, improving site ID & localization in proteomics.




Especially the use of dose-dependent profiling at different time points could clearly separate immediate from late and consequential signaling changes in KRAS-driven (phospho)proteomes.
www.science.org/doi/10.1126/...
(1/4)
Especially the use of dose-dependent profiling at different time points could clearly separate immediate from late and consequential signaling changes in KRAS-driven (phospho)proteomes.
www.nature.com/articles/s41...
www.nature.com/articles/s41...
• (phospho)proteome-wide dose-response profiling
• statistical analysis of 180 million curves with CurveCurator
• mapping kinase-resolved activities changes due to all target engagements
• (re-)evaluating the kinase substrate space in humans
Go #TeamMassSpec!

• (phospho)proteome-wide dose-response profiling
• statistical analysis of 180 million curves with CurveCurator
• mapping kinase-resolved activities changes due to all target engagements
• (re-)evaluating the kinase substrate space in humans
We use zero-distance⚡photo-crosslinking⚡to reveal direct protein-DNA interactions in living cells, enabling quantitative analysis of the DNA-interacting proteome on a timescale of minutes. #DNA #Chromatin #Proteomics
www.cell.com/cell/fulltex...
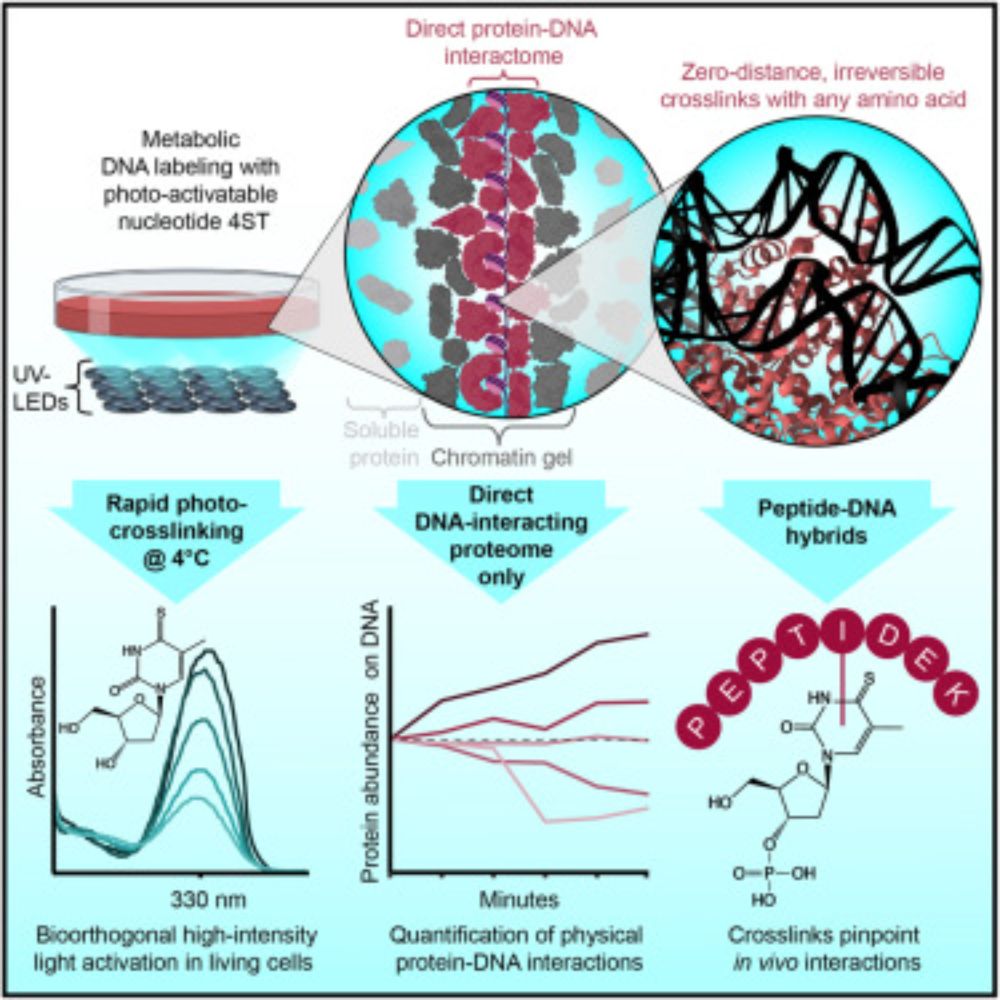
We use zero-distance⚡photo-crosslinking⚡to reveal direct protein-DNA interactions in living cells, enabling quantitative analysis of the DNA-interacting proteome on a timescale of minutes. #DNA #Chromatin #Proteomics
www.cell.com/cell/fulltex...
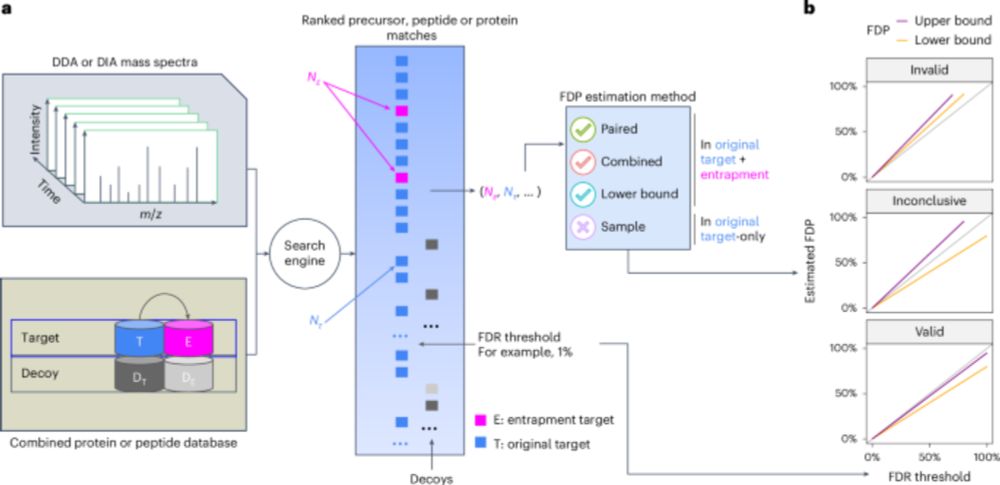
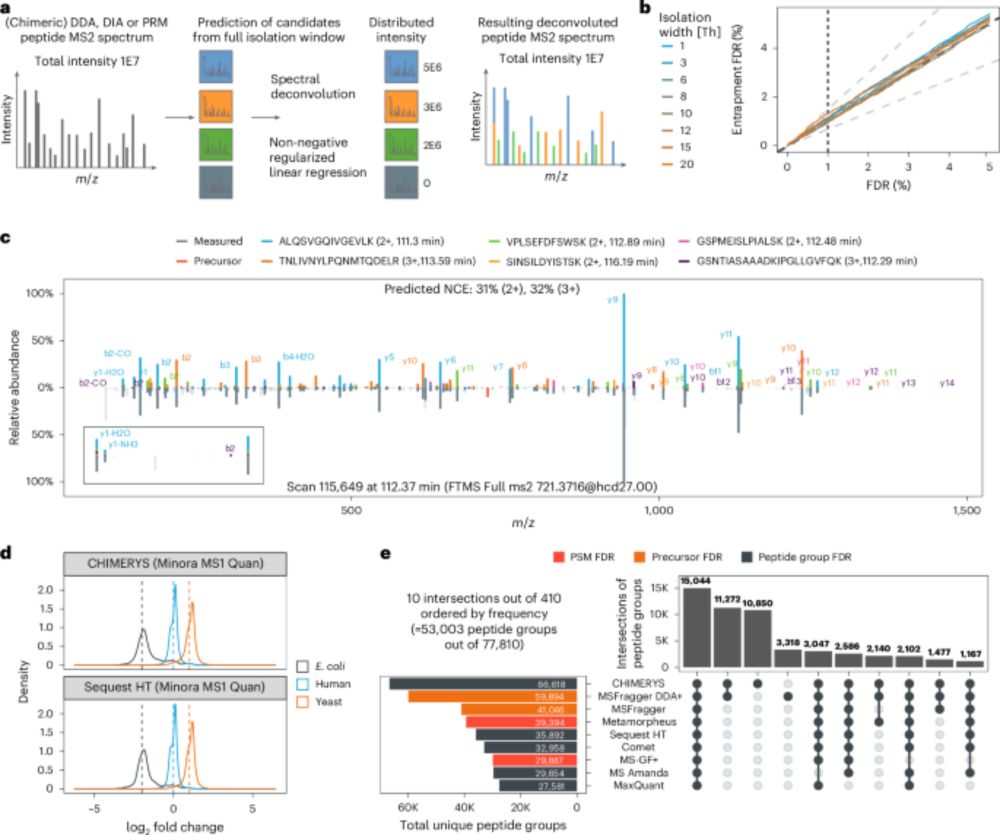
With #CHIMERYS, we can now directly compare DDA and DIA data — 🍎 to 🍎 finally made possible.
doi.org/10.1038/s41592-025-02663-w
#KusterLab #WilhelmLab #MSAID #Proteomics
With #CHIMERYS, we can now directly compare DDA and DIA data — 🍎 to 🍎 finally made possible.
doi.org/10.1038/s41592-025-02663-w
#KusterLab #WilhelmLab #MSAID #Proteomics

There is such a mess in the literature caused by either low throughput or single-dose perturbation experiments. That needs to be solved!
Just scaling up is not enough.
nikolai.slavovlab.net/high-through...

There is such a mess in the literature caused by either low throughput or single-dose perturbation experiments. That needs to be solved!
CurveCurator is the perfect match for fast and reliable statistical analysis of these dose-response data sets.
---
#proteomics #prot-paper

CurveCurator is the perfect match for fast and reliable statistical analysis of these dose-response data sets.
Our dose-resolved combination treatments with (phospho)proteome-wide readouts provide unprecedented quantitative detail of the DNA damage response.
Read more:
www.embopress.org/doi/full/10....

Our dose-resolved combination treatments with (phospho)proteome-wide readouts provide unprecedented quantitative detail of the DNA damage response.
Read more:
www.embopress.org/doi/full/10....
Join us for a celebration on Monday, March 10 !
cos.northeastern.edu/barnett/abou...

Especially decryptM data can be visualized well to see drug potencies for each p-site across a pathway of proteins.
Check it out !!
📄 doi.org/10.1038/s414... (1/6)
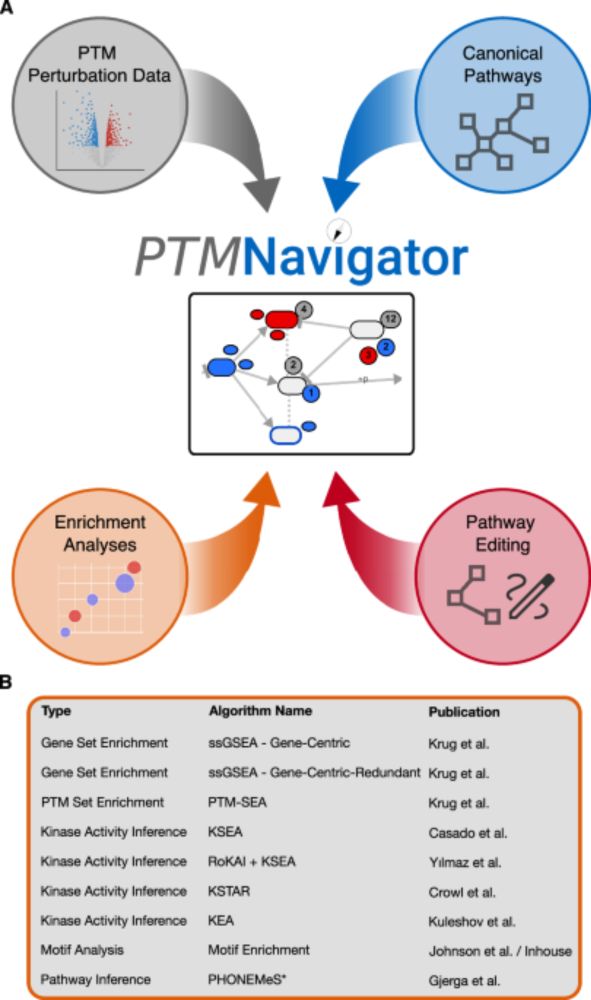
Especially decryptM data can be visualized well to see drug potencies for each p-site across a pathway of proteins.
Check it out !!

