
https://www.uib.no/en/rg/protein

FEBS Workshop Protein Termini 2026: the power of protein termini across bacteria, plants, and animals—from ribosome biology to proteostasis and applications. Deadline: 3 March 2026
proteintermini.org/meeting
#ProteinTermini #Proteostasis #FEBS #EMBO @iubmb.bsky.social

FEBS Workshop Protein Termini 2026: the power of protein termini across bacteria, plants, and animals—from ribosome biology to proteostasis and applications. Deadline: 3 March 2026
proteintermini.org/meeting
#ProteinTermini #Proteostasis #FEBS #EMBO @iubmb.bsky.social
We especially encourage students and postdocs to attend and share their work. All talks (except the keynotes) will be selected from abstracts.
I’m delighted to share that registration is now open for the UK Proteostasis Meeting 2026, hosted by The Francis Crick Institute on 20–21 July 2026 .
Please register here(lnkd.in/ervXMzWN) and through Eventbrite for payment (lnkd.in/eTxqjnQy)

We especially encourage students and postdocs to attend and share their work. All talks (except the keynotes) will be selected from abstracts.
The shining bounds of N-degron pathway influence expands! @charlene-kunaka.bsky.social www.biorxiv.org/content/10.6...
The shining bounds of N-degron pathway influence expands! @charlene-kunaka.bsky.social www.biorxiv.org/content/10.6...
#ProteinTermini #Proteostasis #ProteinModifications #StructuralBiology #PalazzoDeiNormanni #Palermo2026
proteintermini.org/meeting/

#ProteinTermini #Proteostasis #ProteinModifications #StructuralBiology #PalazzoDeiNormanni #Palermo2026
proteintermini.org/meeting/



Check out this one from 1976! Read it here 👉 lnkd.in/egdN-AXt

Check out this one from 1976! Read it here 👉 lnkd.in/egdN-AXt
2. Learn interdisc skills and discover cool new things about how mitochondria move and socialise
3. Explore some of Norway's beautiful nature (both the below, Rundemanen and Gullfjellet, <10km from work)


2. Learn interdisc skills and discover cool new things about how mitochondria move and socialise
3. Explore some of Norway's beautiful nature (both the below, Rundemanen and Gullfjellet, <10km from work)
but the very next day, MetAP took it away.
This year, to save histones from tears,
NatD gives you an acetyl group. ⭐️
Explore our latest paper with the Deuerling lab @uni-konstanz.de and Shu-ou Chan lab @caltech.edu!
www.science.org/doi/10.1126/...

but the very next day, MetAP took it away.
This year, to save histones from tears,
NatD gives you an acetyl group. ⭐️
Explore our latest paper with the Deuerling lab @uni-konstanz.de and Shu-ou Chan lab @caltech.edu!
www.science.org/doi/10.1126/...

🎨 The cover brings warm, summertime vibes ☀️ and highlights a Feature Review on #AromaticAminoAcid biosynthesis in 🌱 from Jorge El-Azaz and @hiroshi-maeda.bsky.social. Cover art credit to Tae Park.
(1/n)

🎨 The cover brings warm, summertime vibes ☀️ and highlights a Feature Review on #AromaticAminoAcid biosynthesis in 🌱 from Jorge El-Azaz and @hiroshi-maeda.bsky.social. Cover art credit to Tae Park.
(1/n)
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
www.conferencecentral.org/webpage/view...
www.conferencecentral.org/webpage/view...
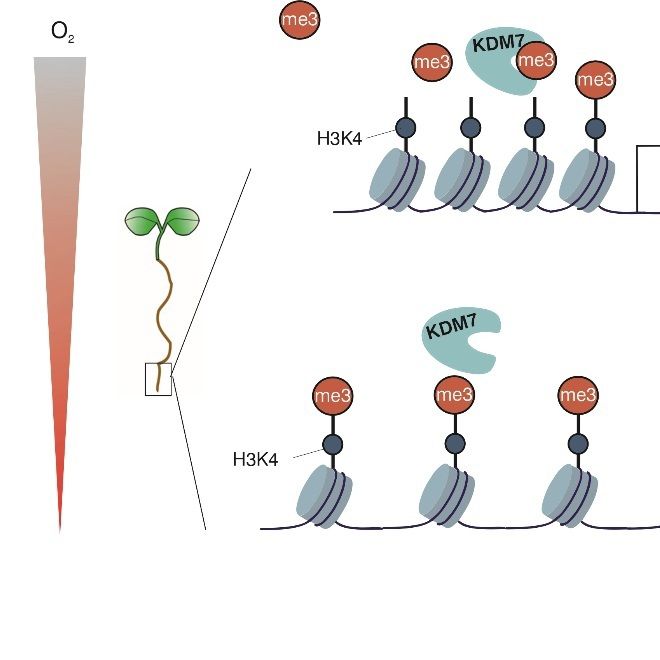
#PostTranslationalModifications #DNADamageResponse #Proteostasis #Autophagy #MetabolicRegulation
Read it here: authors.elsevier.com/a/1m4nM3S6Gf...

#PostTranslationalModifications #DNADamageResponse #Proteostasis #Autophagy #MetabolicRegulation
Read it here: authors.elsevier.com/a/1m4nM3S6Gf...


Lab website: sites.google.com/site/danielg...
Lab website: sites.google.com/site/danielg...

🎨 The cover highlights a Review from @chiosislabmskcc.bsky.social and co on how PTMs reprogram chaperones into epichaperomes.

🎨 The cover highlights a Review from @chiosislabmskcc.bsky.social and co on how PTMs reprogram chaperones into epichaperomes.

