To scale outer membrane protein island simulated at coarse-grained resolution with portions refined at all atom resolution.

To scale outer membrane protein island simulated at coarse-grained resolution with portions refined at all atom resolution.
Stop peddling in unscientific discourse about “AGI” and “superintelligence.” Serve citizens. Don't cater to the whims of tech CEOs.
www.techpolicy.press/ai-hype-is-s...


How do nuclear membranes fuse during NPC assembly? We answer this question in our latest work, where we identify a new mechanism for membrane fusion… (1/13)
How do nuclear membranes fuse during NPC assembly? We answer this question in our latest work, where we identify a new mechanism for membrane fusion… (1/13)
clauswilke.substack.com/p/no-alphafo...
clauswilke.substack.com/p/no-alphafo...
www.aljazeera.com/news/2025/6/...

www.aljazeera.com/news/2025/6/...
➡️ Read more here: bit.ly/42LlJm9
📑 Read the full article: rdcu.be/ef6YX
📝 Support our statement: bit.ly/3zVS3qm
#MDDB #FAIRdata #OpenScience #MolecularDynamics
📰 @lavanguardia.com
🔗 https://f.mtr.cool/ncjierxuzt
#IRBScience #MolecularSimulation
🧪

➡️ Read more here: bit.ly/42LlJm9
📑 Read the full article: rdcu.be/ef6YX
📝 Support our statement: bit.ly/3zVS3qm
#MDDB #FAIRdata #OpenScience #MolecularDynamics
IG 3sixteen
IG 3sixteen

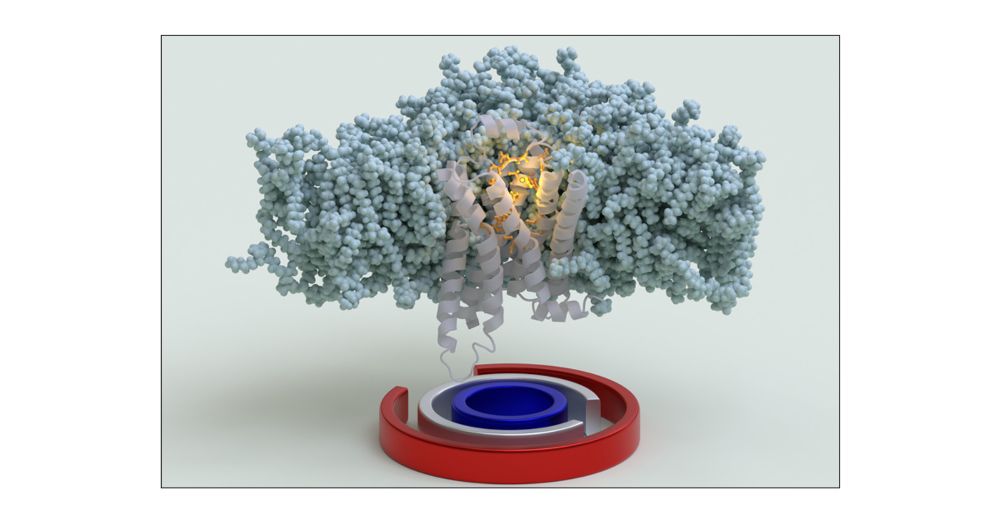
With @saureli.bsky.social @valeriorizzi.bsky.social and Nicola we used OneOPES to fully converge the free energy landscapes associated with B1AR apo/holo activation.
With @saureli.bsky.social @valeriorizzi.bsky.social and Nicola we used OneOPES to fully converge the free energy landscapes associated with B1AR apo/holo activation.
www.embl.org/about/info/e...
www.embl.org/about/info/e...
https://go.nature.com/3QGKmJu

https://go.nature.com/3QGKmJu
BioEmu from MSR: Scalable emulation of protein equilibrium ensembles with generative deep learning www.biorxiv.org/content/10.1...
Looking forward to hearing more explanainers in tomorrow's reading group session on zoom at 9am PT / 12pm ET / 6pm CET: portal.valencelabs.com/logg
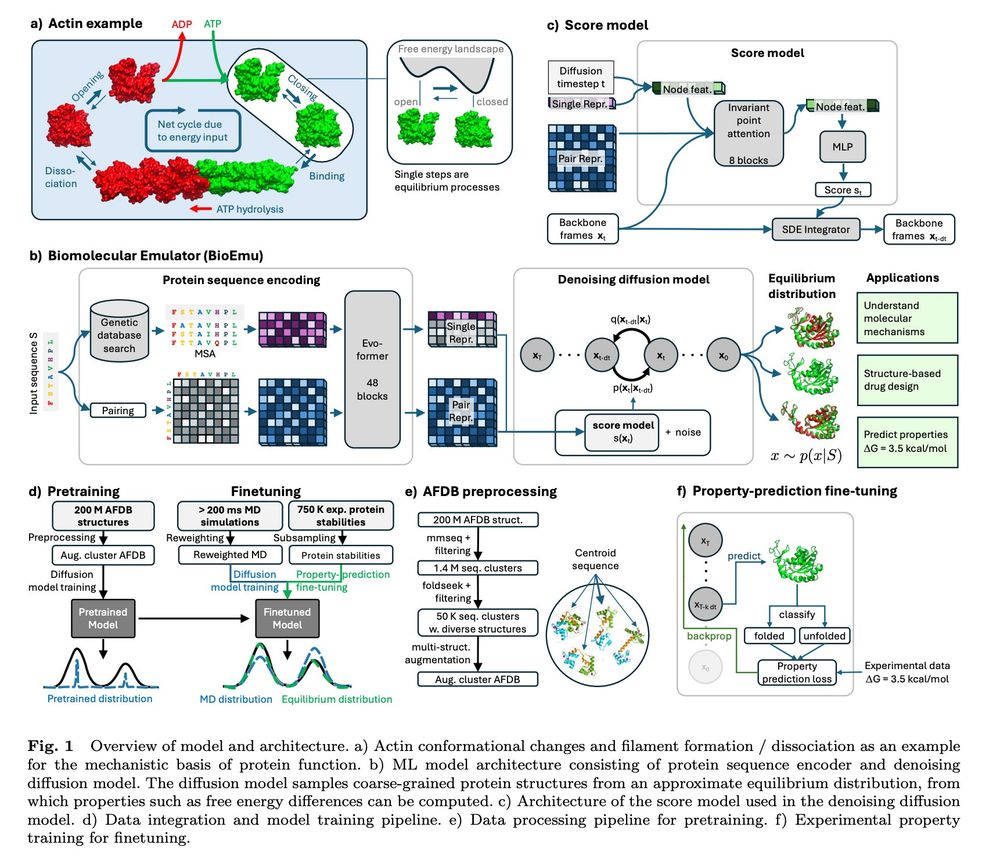
BioEmu from MSR: Scalable emulation of protein equilibrium ensembles with generative deep learning www.biorxiv.org/content/10.1...
Looking forward to hearing more explanainers in tomorrow's reading group session on zoom at 9am PT / 12pm ET / 6pm CET: portal.valencelabs.com/logg
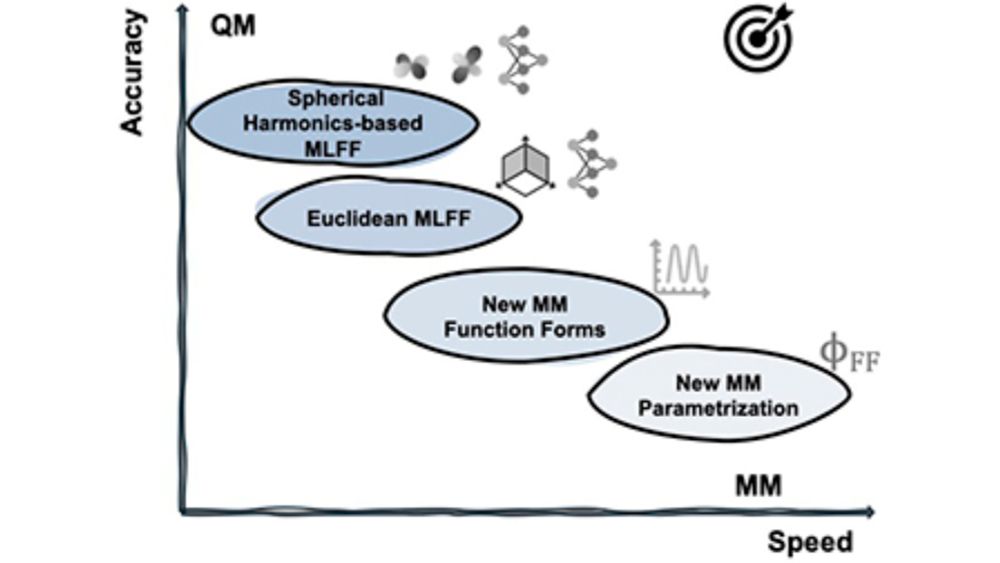
In collaboration with Pande's lab at JnJ and Cournia's lab at BRFAA we reviewed the state of the art in silico tools for studying cryptic pockets.
In collaboration with Pande's lab at JnJ and Cournia's lab at BRFAA we reviewed the state of the art in silico tools for studying cryptic pockets.
bbglab.irbbarcelona.org/2024/12/job-...
#IRBJobs

bbglab.irbbarcelona.org/2024/12/job-...
#IRBJobs

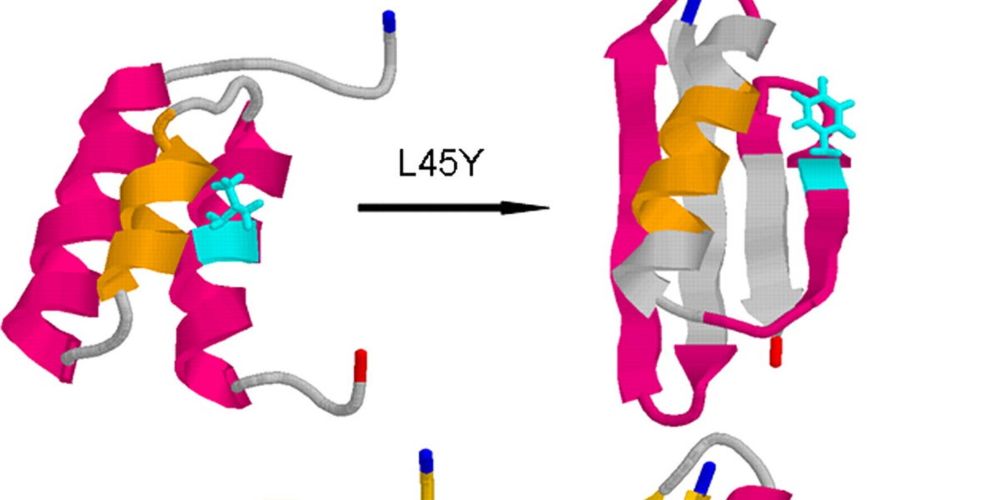
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
biorxiv.org/content/10.1...
Code + Ensembles:
github.com/paulrobustel...
biorxiv.org/content/10.1...
Code + Ensembles:
github.com/paulrobustel...

www.epfl.ch/labs/cosmo/i...

www.epfl.ch/labs/cosmo/i...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...


