We will soon open postdoctoral positions across several components of the project! An expression-of-interest form is already online.

Feedback is welcome. Submission in two weeks.
lnkd.in/d3MCJhEg

Feedback is welcome. Submission in two weeks.
lnkd.in/d3MCJhEg
We will soon open postdoctoral positions across several components of the project! An expression-of-interest form is already online.
We will soon open postdoctoral positions across several components of the project! An expression-of-interest form is already online.





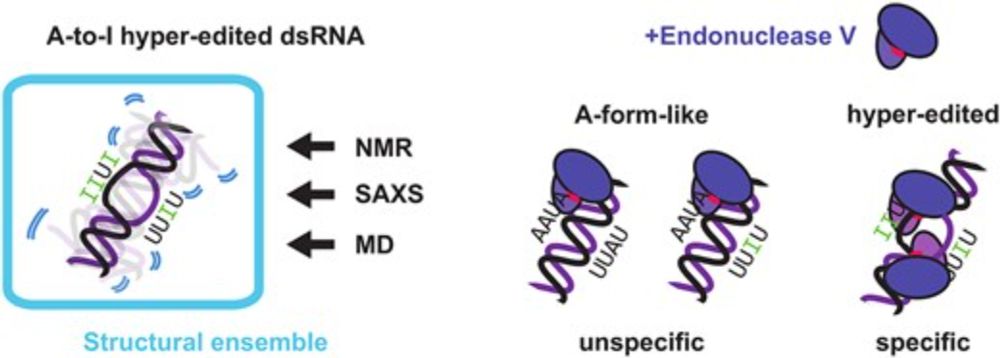
integrate molecular simulations with experimental measurements"!
integrate molecular simulations with experimental measurements"!
arxiv.org/abs/2507.14413
Many thanks, everyone!